Abstract
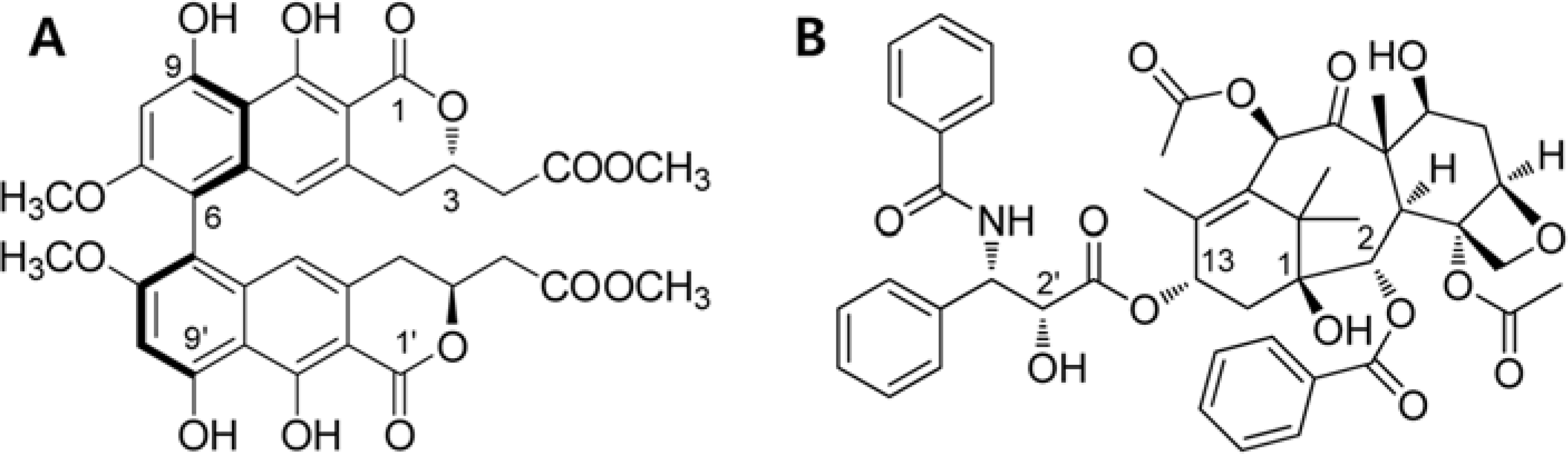
Viriditoxin is a fungal metabolite isolated from Paecilomyces variotii, which was derived from the giant jellyfish Nemopilema nomurai. Viriditoxin was reported to inhibit polymerization of FtsZ, which is a key protein for bacterial cell division and a structural homologue of eukaryotic tubulin. Both tubulin and FtsZ contain a GTP-binding domain, have GTPase activity, assemble into protofilaments, two-dimensional sheets, and protofilament rings, and share substantial structural identities. Accordingly, we hypothesized that viriditoxin may inhibit eukaryotic cell division by inhibiting tubulin polymerization as in the case of bacterial FtsZ inhibition. Docking simulation of viriditoxin to β-tubulin indicated that it binds to the paclitaxel-binding domain and makes hydrogen bonds with Thr276 and Gly370 in the same manner as paclitaxel. Viriditoxin suppressed growth of A549 human lung cancer cells, and inhibited cell division with G2/M cell cycle arrest, leading to apoptotic cell death.
Go to : 
REFERENCES
(1). Liu J., Li F., Kim E. L., Hong J. K., Jung J. H.Nat. Prod. Sci. 2013; 19:61–65.
(2). Wang J., Galgoci A., Kodali S., Herath K. B., Jayasuriya H., Dorso K., Vicente F., González A., Cully D., Bramhill D., Singh S. J.Biol. Chem. 2003; 278:44424–44428.
(3). Wong D. T., Hamill R. L.Biochem. Biophys. Res. Commun. 1976; 71:332–338.
(4). Löwe J., Amos L. A.Nature. 1998; 391:203–206.
(5). Tian W., Xu D., Deng Y. C.Pharmazie. 2012; 67:811–816.
(6). Erickson H. P.Cell. 1995; 80:367–370.
(7). Bi E. F., Lutkenhaus J.Nature. 1991; 354:161–164.
(8). Chen Y., Erickson H. P. J.Biol. Chem. 2005; 280:22549–22554.
(9). Popp D., Iwasa M., Erickson H. P., Narita A., Maéda Y., Robinson R. C. J.Biol. Chem. 2010; 285:11281–11289.
(10). Anderson D. E., Kim M. B., Moore J. T., O'Brien T. E., Sorto N. A., Grove C. I., Lackner L. L., Ames J. B., Shaw J. T.ACS Chem. Biol. 2012; 7:1918–1928.
(11). Läppchen T., Pinas V. A., Hartog A. F., Koomen G. J., Schaffner-Barbero C., Andreu J. M., Trambaiolo D., Löwe J., Juhem A., Popov A. V., den Blaauwen T.Chem. Biol. 2008; 15:189–199.
(12). Foss M. H., Eun Y. J., Grove C. I., Pauw D. A., Sorto N. A., Rensvold J. W., Pagliarini D. J., Shaw J. T., Weibel D. B.Med. Chem. Commun. 2013; 4:112–119.
(13). Gupta K. K., Bharne S. S., Rathinasamy K., Naik N. R., Panda D.FEBS J. 2006; 273:5320–5332.
(14). Rai D., Singh J. K., Roy N., Panda D.Biochem. J. 2008; 410:147–155.
(15). Wang J., Galgoci A., Kodali S., Herath K. B., Jayasuriya H., Dorso K., Vicente F., González A., Cully D., Bramhill D., Singh S. J.Biol. Chem. 2003; 278:44424–44428.
(16). Hsiao C. J., Hsiao G., Chen W. L., Wang S. W., Chiang C. P., Liu L. Y., Guh J. H., Lee T. H., Chung C. L. J.Nat. Prod. 2014; 77:758–765.
(17). Zhu Z., Sun H., Ma G., Wang Z., Li E., Liu Y., Liu Y.Int. J. Mol. Sci. 2012; 13:2025–2035.
(18). Kundu S., Kim T. H., Yoon J. H., Shin H. S., Lee J., Jung J. H., Kim H. S.Int. J. Oncol. 2014; 45:2331–2340.
(19). Löwe J., Li H., Downing K. H., Nogales E. J.Mol. Biol. 2001; 313:1045–1057.
(20). Sun L., Simmerling C., Ojima I.ChemMedChem. 2009; 4:719–731.
Go to : 
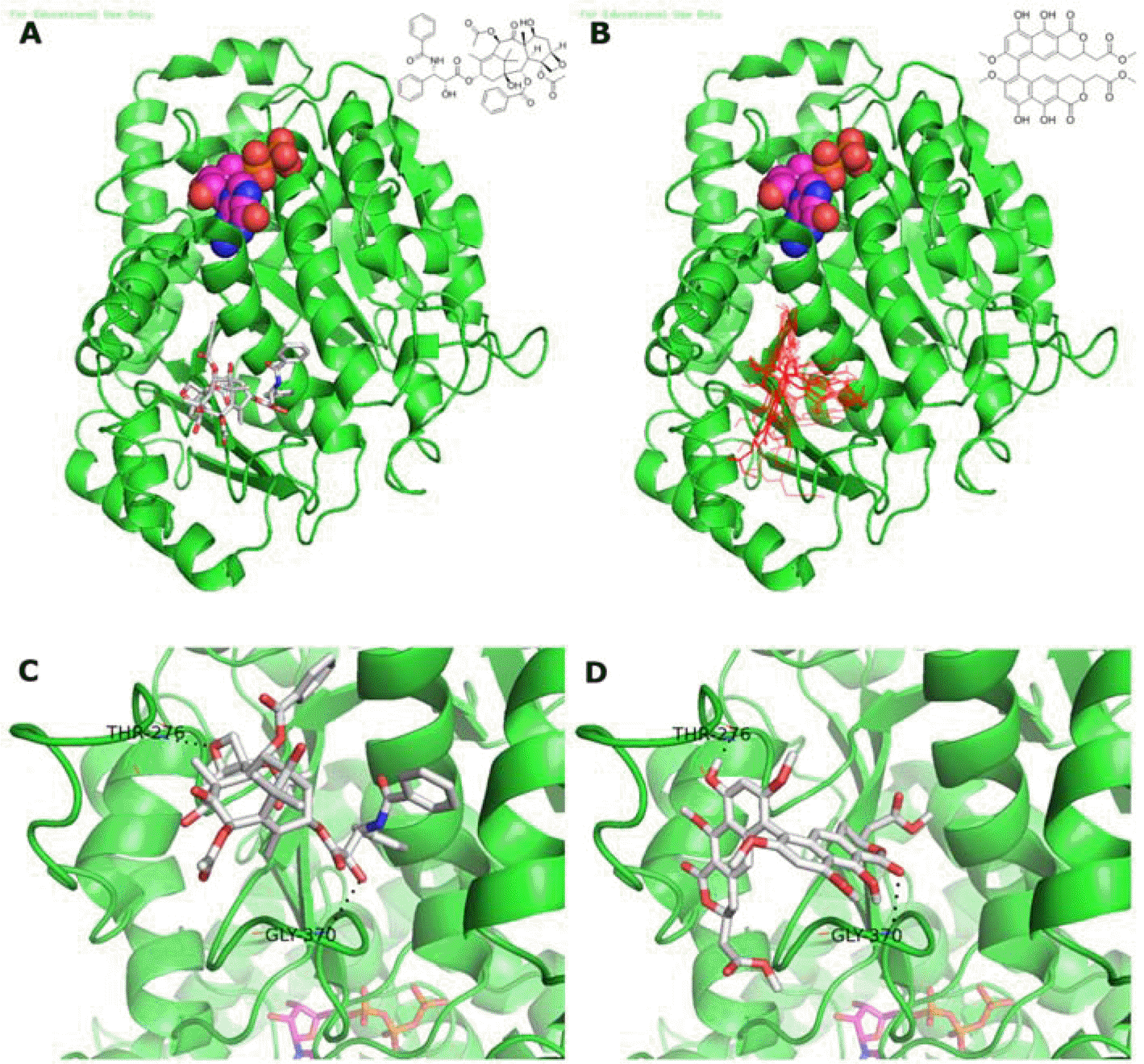 | Fig. 2.Binding modes of viriditoxin and paclitaxel to the ligand binding domain of β-tubulin. GDP is presented as ball and stick. (A) Co-crystal structure of paclitaxel bound to β-tubulin (PDB code number 1JFF). (B) Docking results of viriditoxin to β-tubulin. Viriditoxin binds to the paclitaxel binding domain. (C) Paclitaxel interaction with loop of β-tubulin. Oxetane of paclitaxel makes a hydrogen bond with Thr276. The C-2′ hydroxyl group of paclitaxel makes hydrogen bond with the backbone NH of Gly370. (D) Model of viriditoxin which makes hydrogen bonds with Gly370 and Thr276. The C-9′ hydroxyl group of viriditoxin makes hydrogen bond with Thr276. The C-1 carbonyl group of α-pyrone of viriditoxin makes hydrogen bond with the backbone NH of Gly370. |
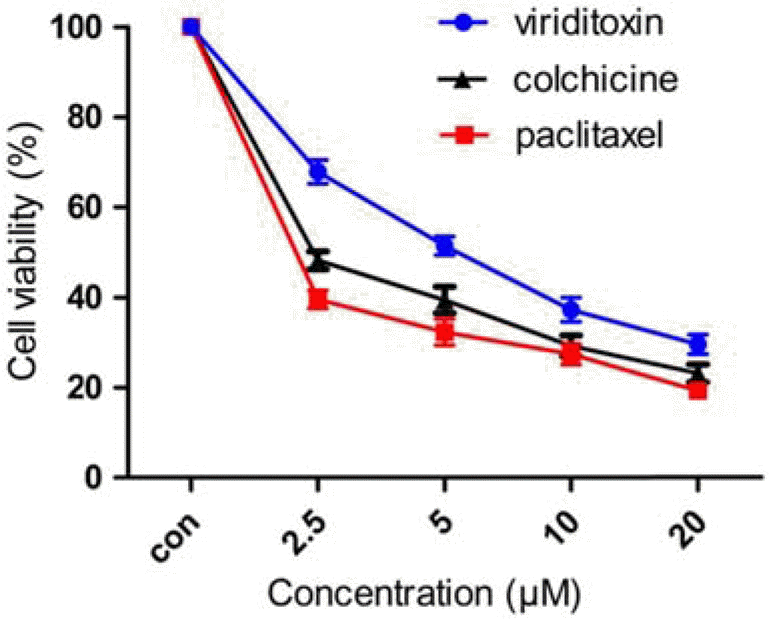 | Fig. 3.Effect of viriditoxin, paclitaxel, and colchicine on viability of A549 cells. The cells were treated with viriditoxin, paclitaxel, or colchicine at various concentrations (0-20 μ M) for 24 h. The percentage of cell survival was determined as the ratio between treated cells and untreated controls. Results are expressed as mean ± S.E.M., n=3. |
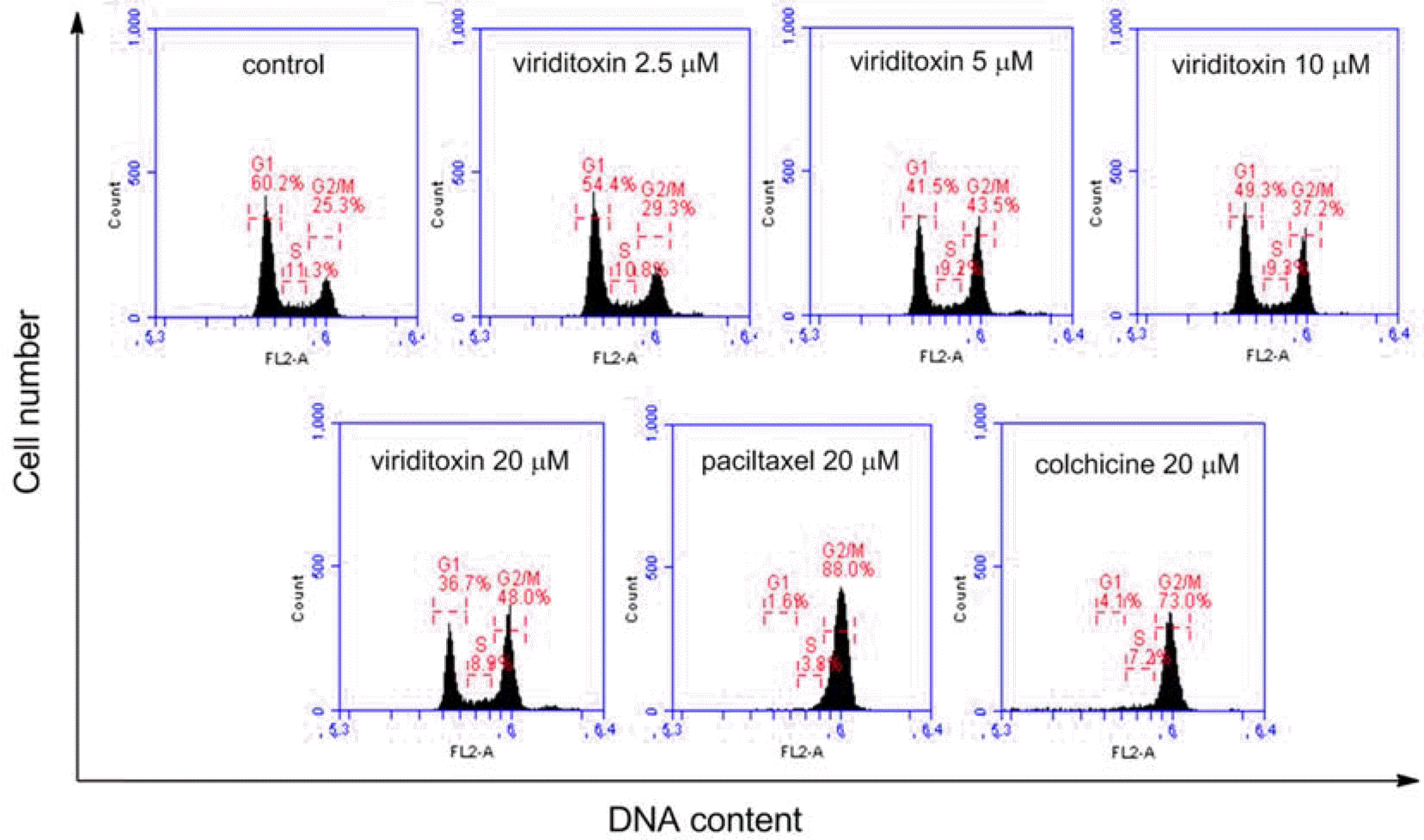 | Fig. 4.Cell cycle analysis of A549 cells treated with viriditoxin, paclitaxel, or colchicine. A549 cells were treated for 24 h with viriditoxin (0-20 μM), paclitaxel (20 μM) or colchicine (20 μM). The cells were fixed and digested with RNase, and then cellular DNA was stained with PI and analyzed by flow cytometry. The percentages of cells in G0/G1, S, and G2/M phases of cell cycle were shown. |
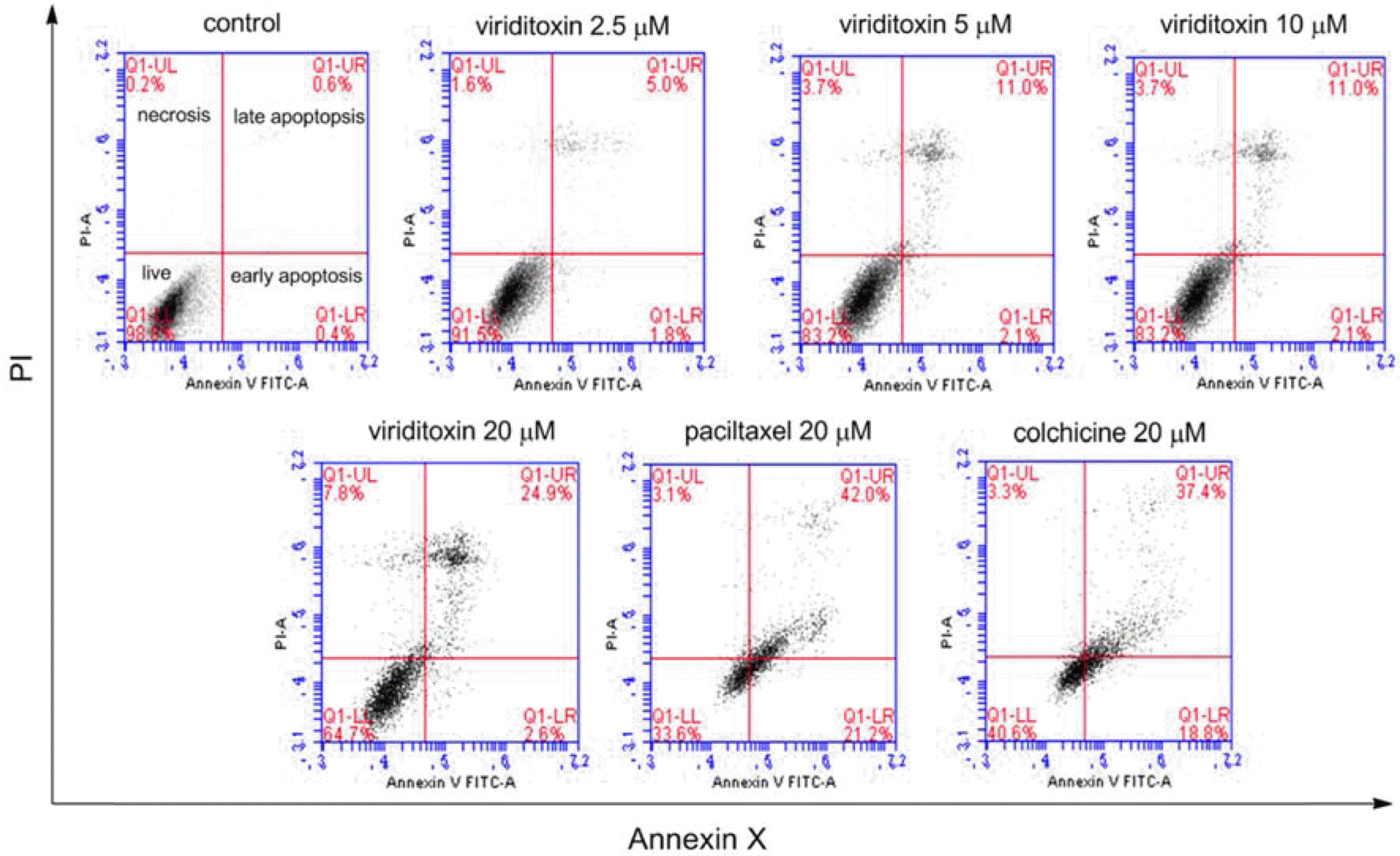 | Fig. 5.Effect of viriditoxin, paclitaxel, and colchicine on apoptosis of A549 cells. Viriditoxin-induced apoptosis in A549 cells was examined by Annexin V-PI binding assay. A549 cells were treated with viriditoxin, paclitaxel, or colchicine for 24 h at different concentrations as indicated. Representative flow cytometry scatter plots depict percent Annexin V/PI staining at 24 h after viriditoxin treatment. Apoptotic cell death was analyzed using flow cytometry. |
Table 1.
The IC50 (μ M) values of viriditoxin and mitotoxins on A549 human lung cancer cells
| Viriditoxin | Colchicine | Paclitaxel |
|---|---|---|
| 5.1 ± 0.4 | 2.3 ± 0.5 | 1.9 ± 0.3 |
Table 2.
Effect of viriditoxin and mitotoxins on cell cycle distribution in A549 cells
Table 3.
Effect of viriditoxin and mitotoxins on apoptosis in A549 cells




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download