Abstract
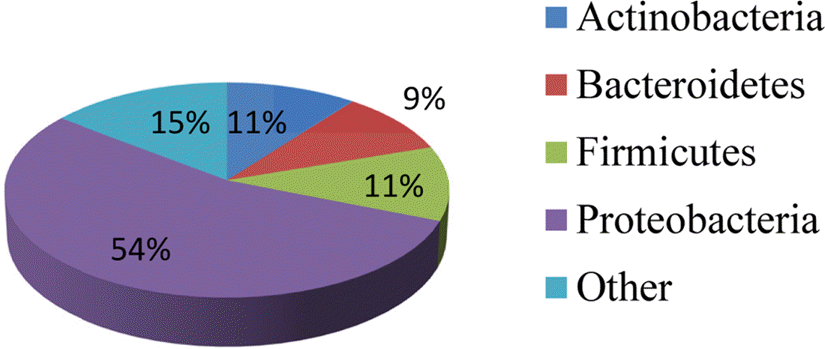
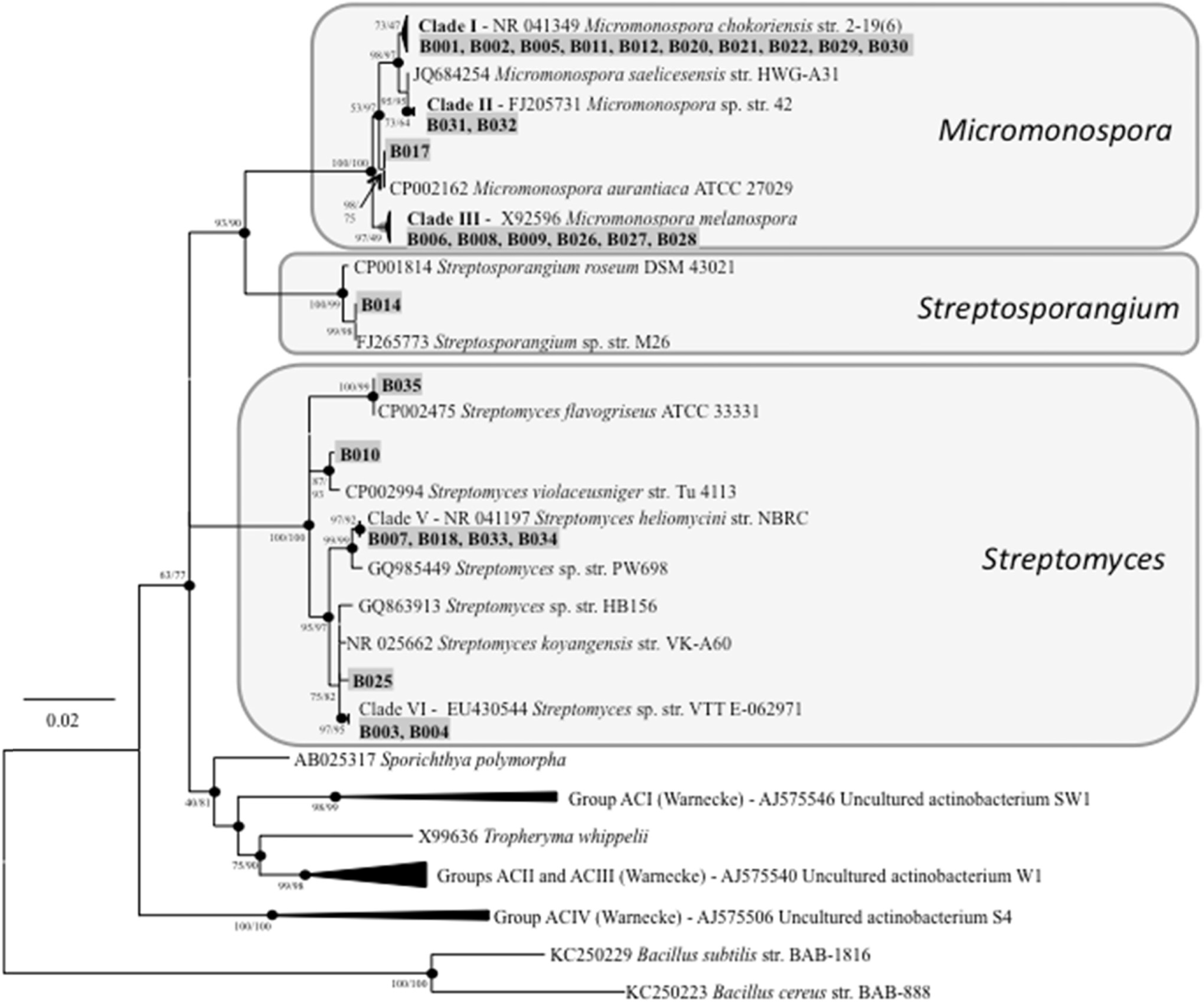
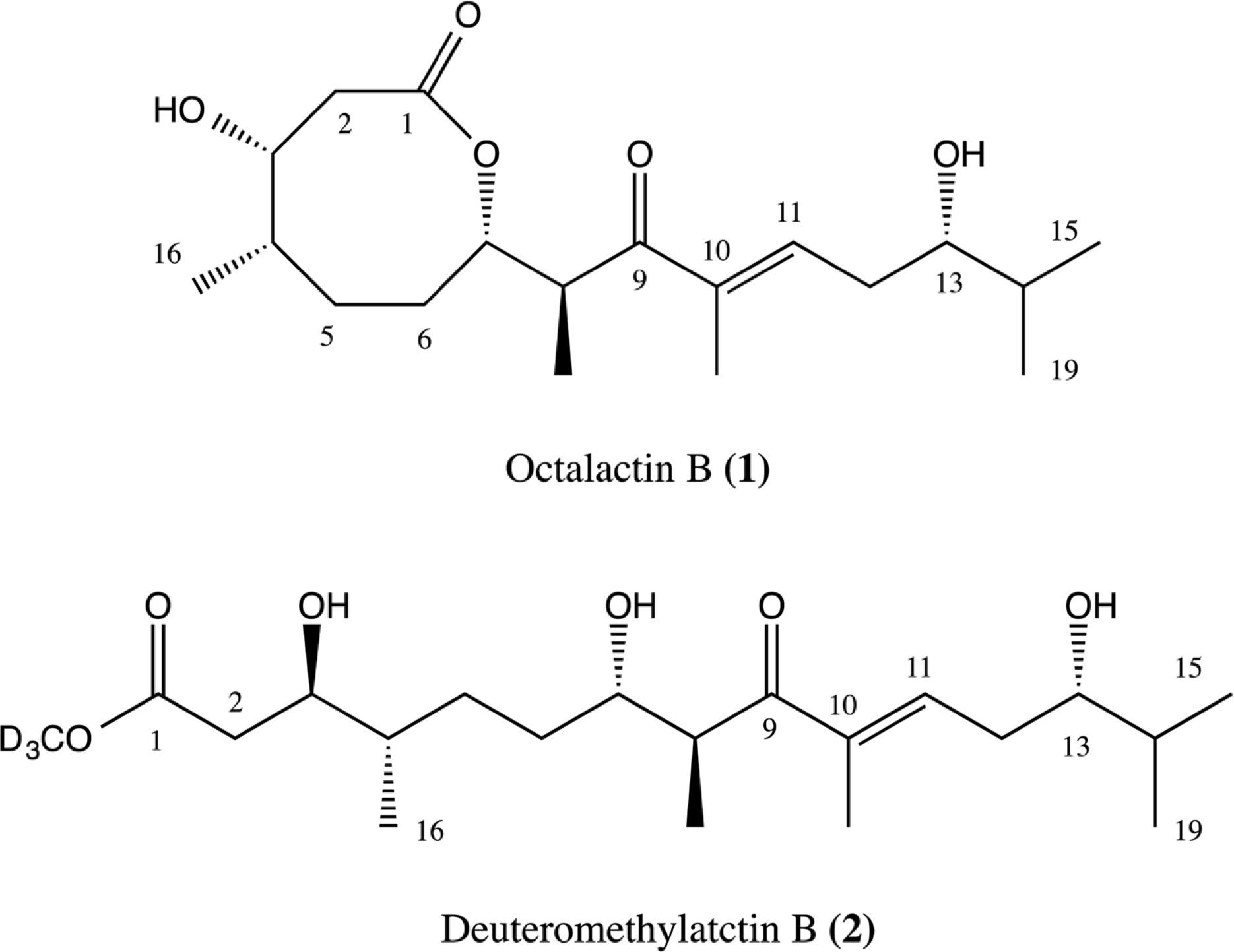
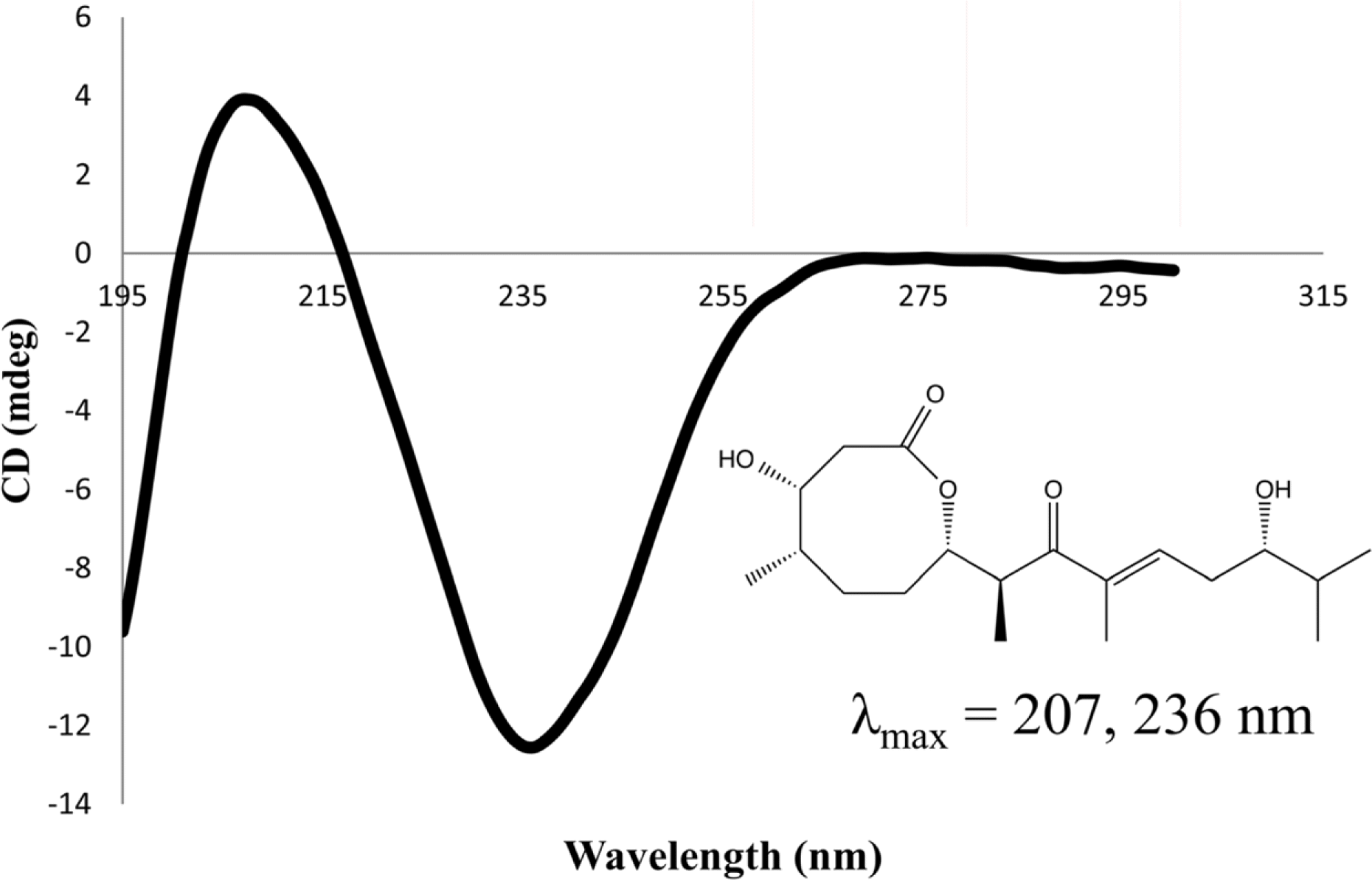
Compared to their terrestrial and marine counterparts, little is known about the capacity of freshwater-derived actinomycete bacteria to produce novel secondary metabolites. In the current study, we highlight the disparities that exist between cultivation-independent and -dependent analyses of actinomycete communities from four locations in Lake Michigan sediment. Furthermore, through phylogenetic analysis of strains isolated from these locations, we identified a Streptomyces sp., strain B025, as being distinct from other Streptomyces spp. isolated from sediment. Upon fermentation this strain produced a rare class of eight-membered lactone secondary metabolites, which have been for their antitumor properties. We used spectroscopic and chemical derivitization techniques to characterize octalactin B (1) in addition to its corresponding novel, unnatural degradation product (2).
Go to : 
REFERENCES
(1). Newman D. J., Cragg G. M. J.Nat. Prod. 2012; 75:311–335.
(2). Fenical W., Jensen P. R.Nat. Chem. Biol. 2006; 2:666–673.
(3). Gerwick W. H., Moore B. S.Chem. Biol. 2012; 19:85–98.
(4). Mullowney M. W., Hwang C. H., Newsome A. G., Wei X., Tanouye U., Wan B., Carlson S., Barranis N. J., ÓhAinmhire E., Chen W. L., Krishnamoorthy K., White J., Blair R., Lee H., Burdette J. E., Rathod P. K., Parish T., Cho S., Franzblau S. G., Murphy B. T.ACS Infect. Dis. 2015; 1:168–174.
(5). Carlson S., Tanouye U., Omarsdottir S., Murphy B. T. J.Nat. Prod. 2015; 78:381–387.
(6). Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., Knight R.ISME J. 2012; 6:1621–1624.
(7). Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R.Nat. Methods. 2010; 7:335–336.
(8). Edgar R. C.Bioinformatics. 2010; 26:2460–2461.
(9). Gihring T. M., Green S. J., Schadt C. W.Environ. Microbiol. 2012; 14:285–290.
(10). McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., Hugenholtz P.ISME J. 2012; 6:610–618.
(11). Wang Q., Garrity G. M., Tiedje J. M., Cole J. R.Appl. Environ. Microbiol. 2007; 73:5261–5267.
(12). Clarke K. R.Aust. J. Ecol. 1993; 18:117–143.
(13). Green S. J., Prakash O., Jasrotia P., Overholt W. A., Cardenas E., Hubbard D., Tiedje J. M., Watson D. B., Schadt C. W., Brooks S. C., Kostka J. E.Appl. Environ. Microbiol. 2012; 78:1039–1047.
(14). DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L.Appl. Environ. Microbiol. 2006; 72:5069–5072.
(15). Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar Buchner A., Lai T., Steppi S., Jobb G., Forster W., Brettske I., Gerber S., Ginhart A. W., Gross O., Grumann S., Hermann S., Jost R., Konig A., Liss T., Lüssmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., Schleifer K. H.Nucleic Acids Res. 2004; 32:1363–1371.
(16). Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.Mol. Biol. Evol. 2011; 28:2731–2739.
(17). Ronquist F., Huelsenbeck J. P.Bioinformatics. 2003; 19:1572–1574.
(18). Jensen P. R., Moore B. S., Fenical W.Nat. Prod. Rep. 2015; 32:738–751.
(19). Newton R. J., Jones S. E., Eiler A., McMahon K. D., Bertilsson S.Microbiol. Mol. Biol. Rev. 2011; 75:14–49.
(20). Glöckner F. O., Zaichikov E., Belkova N., Denissova L., Pernthaler J., Pernthaler A., Amann R.Appl. Environ. Microbiol. 2000; 66:5053–5065.
(21). Zwart G., Crump B. C., Kamst-van Agterveld M. P. K. V., Hagen F., Han S. K.Aquat. Microb. Ecol. 2002; 28:141–155.
(22). Warnecke F., Amann R., Pernthaler J.Environ. Microbiol. 2004; 6:242–253.
(23). Newton R. J., Jones S. E., Helmus M. R., McMahon K. D.Appl. Environ. Microbiol. 2007; 73:7169–7176.
(24). Hahn M. W., Lünsdorf H., Wu Q., Schauer M., Höfle M. G., Boenigk J., Stadler P.Appl. Environ. Microbiol. 2003; 69:1442–1451.
(26). Tapiolas D. M., Roman M., Fenical W., Stout T. J., Clardy J. J.Am. Chem. Soc. 1991; 113:4682–4683.
(27). Dinh M. T., Bouzbouz S., Péglion J. L., Cossy J.Tetrahedron. 2008; 64:5703–5710.
(28). O'Sullivan P. T., Buhr W., Fuhry M. A. M., Harrison J. R., Davies J. E., Feeder N., Marshall D. R., Burton J. W., Holmes A. B. J.Am. Chem. Soc. 2004; 126:2194–2207.
(29). Shiina I., Hashizume M., Yamai Y. S., Oshiumi H., Shimazaki T., Takasuna Y. J., Ibuka R.Chemistry. 2005; 11:6601–6608.
(30). Dinh M. T., BouzBouz S., Peglion J. L., Cossy J.Synlett. 2005; 18:2851–2853.
(31). Shiina I., Oshiumi H., Hashizume M., Yamai Y. S., Ibuka R.Tetrahedron Lett. 2004; 45:543–547.
(32). Radosevich A. T., Chan V. S., Shih H. W., Toste F. D.Angew. Chem. Int. Ed Engl. 2008; 47:3755–3758.
(33). Hoye T. R., Jeffrey C. S., Shao F.Nat. Protoc. 2007; 2:2451–2458.
(34). Sullivan G. R., Dale J. A., Mosher H. S. J.Org. Chem. 1973; 38:2143–2147.
(35). Lee J. Y., Lee J. Y., Jung H. W., Hwang B. K.Int. J. Syst. Evol. Microbiol. 2005; 55:257–262.
Go to : 
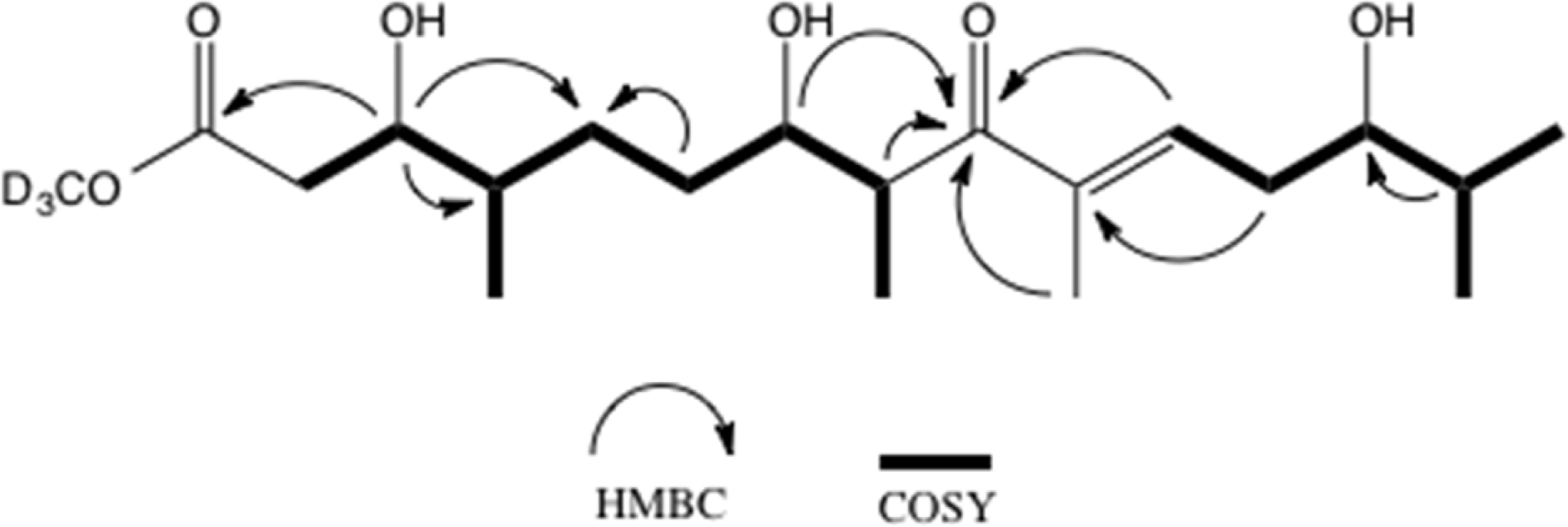
Table 1.
1H and 13C NMR data of 2.
| Position | 13Ca,b | 13Ca,c | 1H mult. (J, Hz)b,d |
|---|---|---|---|
| -OCD3 | 49.0 | 52.0 | |
| 1 | 173.9 | 174.2 | |
| 2 | 39.8 | 38.1 | 2.36 dd (15.1, 9.4) |
| 2.51 dd (15.1, 3.4) | |||
| 3 | 73.4 | 72.7 | 3.88 ddd (9.4, 5.2, 3.4) |
| 4 | 39.9 | 38.3 | 1.55 m |
| 5 | 28.9 | 28.6 | 1.32 m, 1.54 m |
| 6 | 32.5 | 32.7 | 1.32 m, 1.48 m |
| 7 | 74.7 | 74.6 | 3.74 ddd (3.0, 7.3, 8.0) |
| 8 | 46.2 | 43.4 | 3.46 p (7.3) |
| 9 | 207.7 | 207.6 | |
| 10 | 139.4 | 138.5 | |
| 11 | 142.5 | 141.2 | 6.91 t (7.2) |
| 12 | 35.2 | 34.1 | 2.48 m |
| 2.40 dd (16.0, 7.2) | |||
| 13 | 76.7 | 76.0 | 3.49 m |
| 14 | 35.0 | 34.1 | 1.70 m |
| 15 | 19.3 | 18.9 | 0.96 d (7.0) |
| 16 | 15.4 | 15.3 | 0.90 d (6.7) |
| 17 | 14.8 | 16.3 | 1.02 d (7.3) |
| 18 | 11.9 | 11.7 | 1.79 s |
| 19 | 18.0 | 17.6 | 0.97 d (7.0) |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download