Abstract
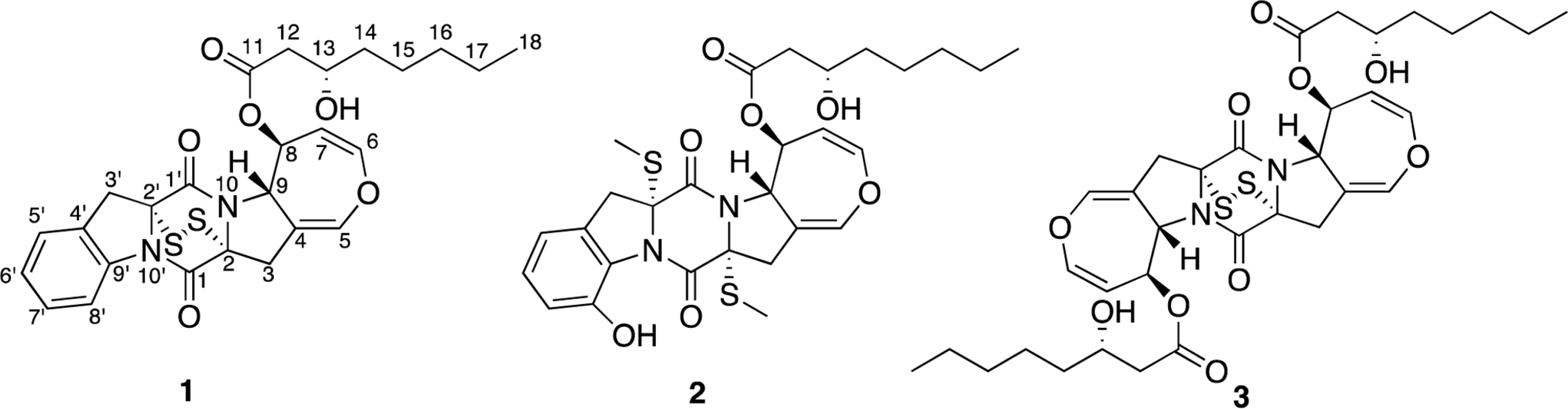
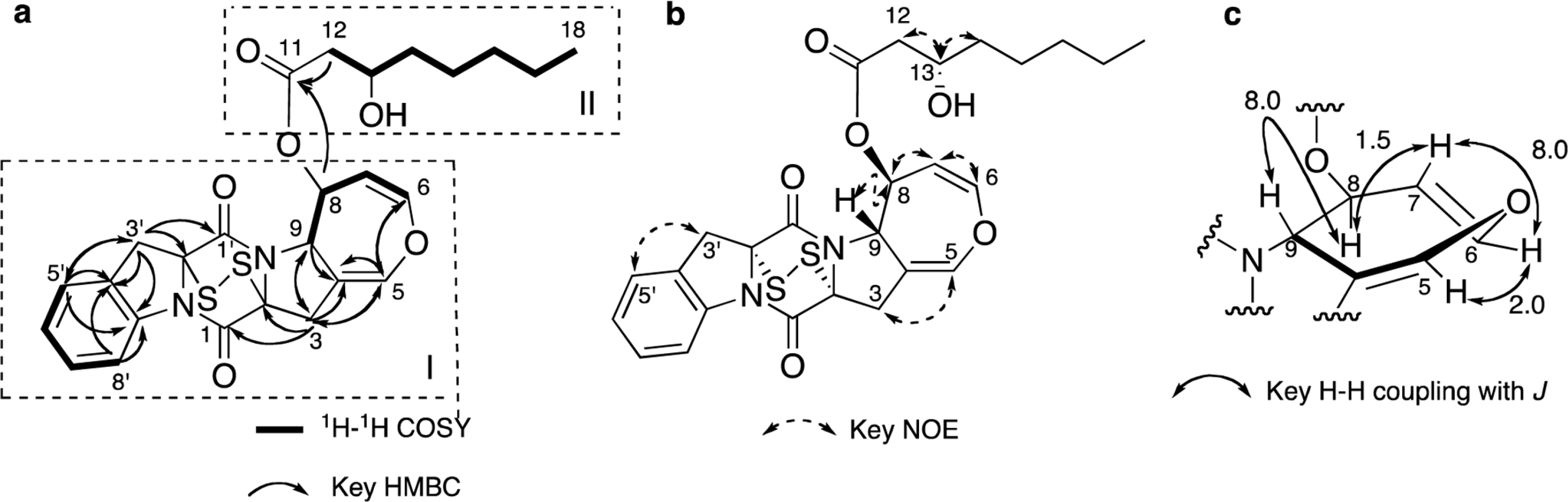
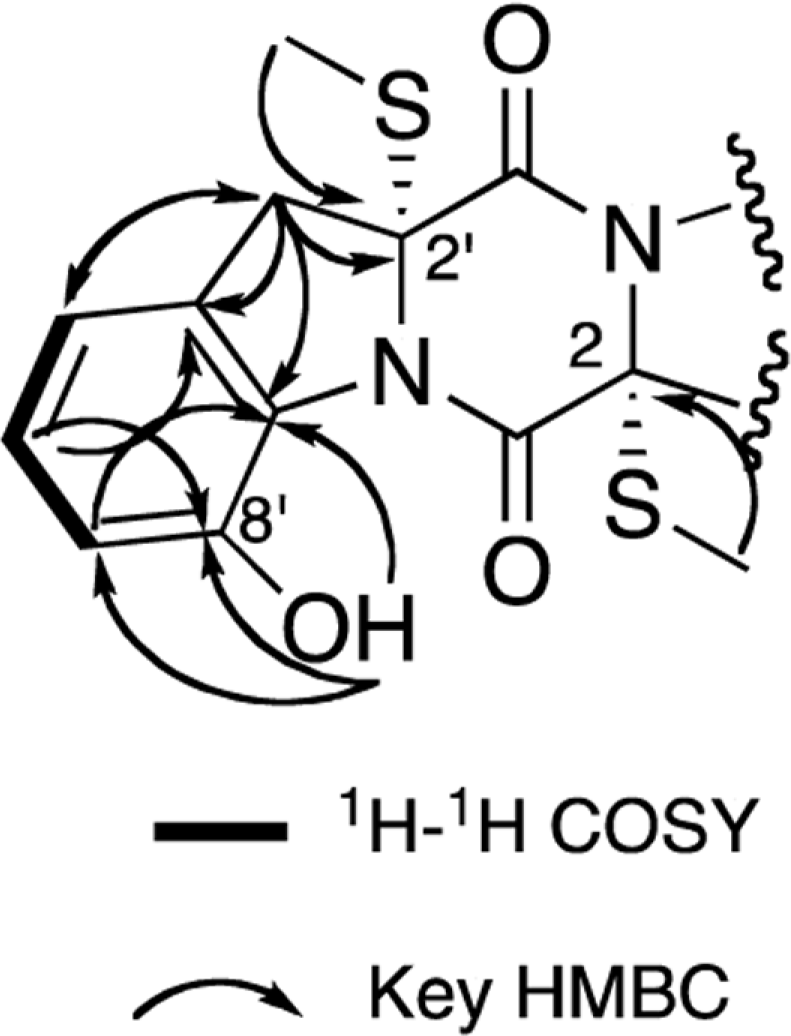
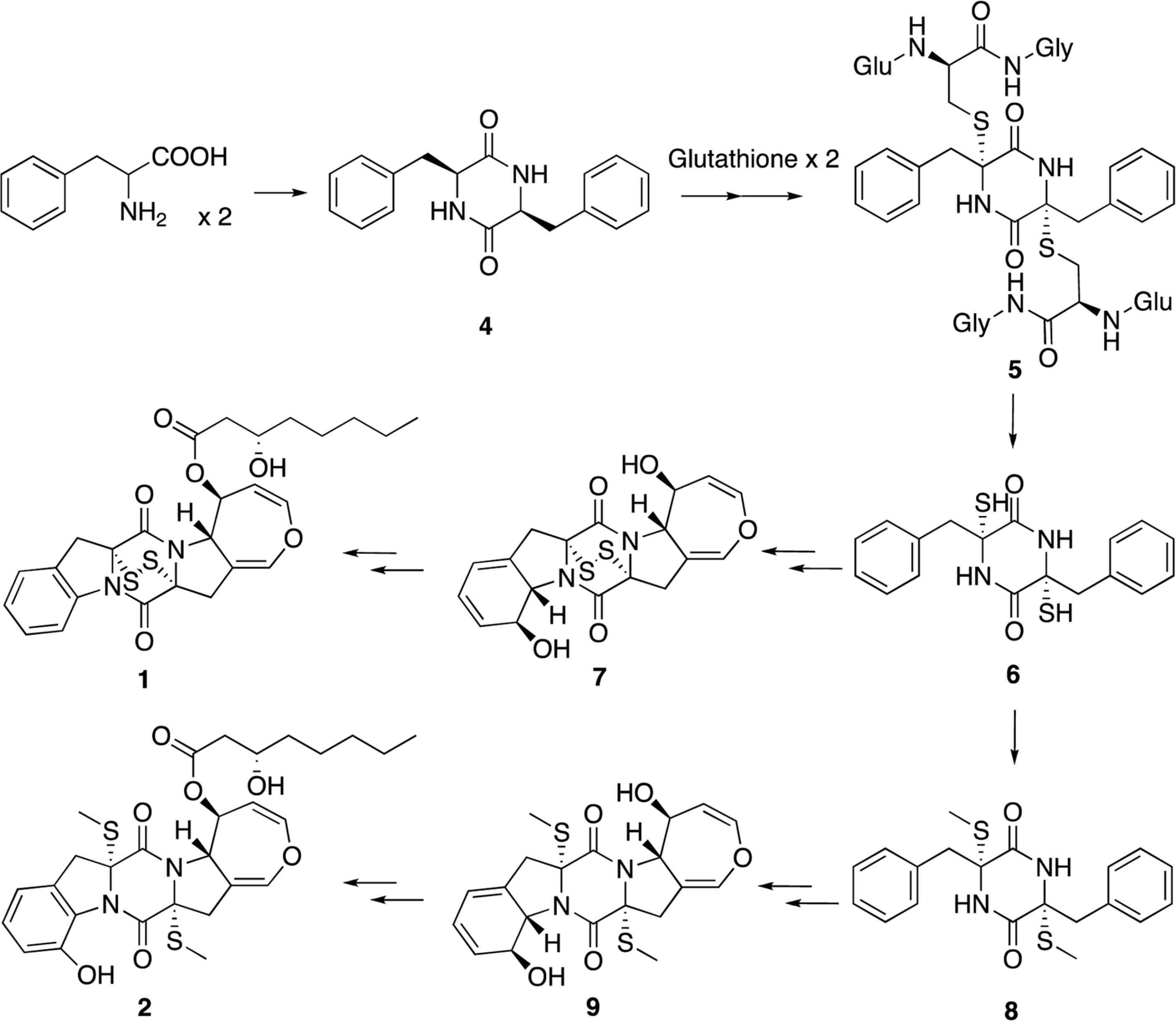
Two new thiodiketopiperazines (TDKPs), designated graphiumins I (1) and J (2), were isolated from the culture broth of the marine-derived fungus Graphium sp. OPMF00224 by solvent extraction, silica gel column chromatography, and HPLC. Their absolute structures were elucidated by spectroscopic analyses (1D and 2D NMR data, ROESY correlations, and CD data) and chemical methods. They were found to be structurally rare TDKPs with a phenylalanine-derived indolin substructure. Compounds 1 and 2 inhibited yellow pigment production by methicillin-resistant Staphylococcus aureus (MRSA) with IC50 values of 63.5 and 76.5 µg/ml, respectively, without inhibiting its growth, even at 250 µ g/ml.
REFERENCES
(1). Centers for Disease Control and Prevention (CDC). MMWR Morb. Mortal. Wkly. Rep. 1997; 46:765–766.
(2). Hiramatsu K., Hanaki H., Ino T., Yabuta K., Oguri T., Tenover F. C. J.Antimicrob. Chemother. 1997; 40:135–136.
(3). Marshall J. H., Wilmoth G. J. J.Bacteriol. 1981; 147:900–913.
(4). Marshall J. H., Wilmoth G. J. J.Bacteriol. 1981; 147:914–919.
(5). Clauditz A., Resch A., Wieland K. P., Peschel A., Götz F.Infect. Immun. 2006; 74:4950–4953.
(6). Liu G. Y., Essex A., Buchanan J. T., Datta V., Hoffman H. M., Bastian J. F., Fierer J., Nizet V. J.Exp. Med. 2005; 202:209–215.
(7). Liu C. I., Liu G. Y., Song Y., Yin F., Hensler M. E., Jeng W. Y., Nizet V., Wang A. H., Oldfield E.Science. 2008; 319:1391–1394.
(8). Song Y., Liu C. I., Lin F. Y., No J. H., Hensler M. E., Liu Y. L., Jeng W. Y., Low J., Liu G. Y., Nizet V., Wang A. H., Oldfield E. J.Med. Chem. 2009; 52:3869–3880.
(9). Liu C. I., Jeng W. Y., Chang W. J., Ko T. P., Wang A. H. J.Biol. Chem. 2012; 287:18750–18757.
(10). Lee J. H., Cho H. S., Kim Y., Kim J. A., Banskota S., Cho M. H., Lee J.Appl. Microbiol. Biotechnol. 2013; 97:4543–4552.
(11). Lee J. H., Park J. H., Cho M. H., Lee J.Curr. Microbiol. 2012; 65:726–732.
(12). Sakai K., Koyama N., Fukuda T., Mori Y., Onaka H., Tomoda H.Biol. Pharm. Bull. 2012; 35:48–53.
(13). Fukuda T., Nagai K., Tomoda H. J.Nat. Prod. 2012; 75:2228–2231.
(14). Fukuda T., Shimoyama K., Nagamitsu T., Tomoda H. J.Antibiot. 2014; 67:445–450.
(15). Fukuda T., Shinkai M., Sasaki E., Nagai K., Kurihara Y., Kanamoto A., Tomoda H. J.Antibiot. 2015; DOI: doi: 10.1038/ja.2015.41.
(16). Wang J. M., Jiang N., Ma J., Yu S. S., Tan R. X., Dai J. G., Si Y. K., Ding G. Z., Ma S. G., Qu J., Fang L., Du D.Tetrahedron. 2013; 69:1195–1201.
(17). Nagai K., Doi T., Sekiguchi T., Namatame I., Sunazuka T., Tomoda H., Omura S., Takahashi T. J.Comb. Chem. 2006; 8:103–109.
(18). Hegde V. R., Dai P., Patel M., Das P. R., Puar M. S.Tetrahedron Lett. 1997; 38:911–914.
(19). Neuss N., Nagarajan R., Molloy B. B., Huckstep L. L.Tetrahedron Lett. 1968; 9:4467–4471.
(20). Guo C. J., Yeh H. H., Chiang Y. M., Sanchez J. F., Chang S. L., Bruno K. S., Wang C. C. J.Am. Chem. Soc. 2013; 135:7205–7213.
Table 1.
13C and 1H NMR spectroscopic data for graphiumins I (1) and J (2) in CDCl3
| 1 | 2 | |||
|---|---|---|---|---|
| Position | δCa | δH mult (J in Hz)b | δCa | δH mult (J in Hz)b |
| 1 | 160.4, s | 169.2, s | ||
| 2 | 75.7, s | 69.9, s | ||
| 3 | 34.2, t | 4.15, dq (18.0, 1.0) | 39.3, t | 3.23, dt (17.0, 1.0) |
| 3.10, dq (18.0, 1.5) | 3.13, dt (17.0, 1.5) | |||
| 4 | 113.1, s | 108.9, s | ||
| 5 | 139.7, d | 6.69, q (2.0) | 138.2, d | 6.64, t (2.0) |
| 6 | 141.6, d | 6.35, dd (8.0, 2.0) | 139.9, d | 6.34, dd (8.0, 2.0) |
| 7 | 105.0, d | 4.61, dd (8.0, 1.5) | 105.5, d | 4.69, dd (8.0, 2.0) |
| 8 | 69.9, d | 5.86, dt (8.0, 1.5) | 71.9, d | 5.93, dt (8.5, 2.0) |
| 9 | 63.0, d | 5.18, dq (8.0, 1.5) | 60.5, d | 5.21, br d (8.5) |
| 10 | ||||
| 11 | 171.7, s | 171.6, s | ||
| 12 | 41.9, t | 2.56, dd (17.0, 3.0) | 42.1, t | 2.56, dd (17.0, 3.0) |
| 2.46, dd (17.0, 9.0) | 2.47, dd (17.0, 9.0) | |||
| 13 | 67.6, d | 4.09, m | 67.5, d | 4.07, m |
| 14 | 36.5, t | 1.44, m | 36.5, t | 1.43, m |
| 1.52, m | 1.51, m | |||
| 15 | 25.3, t | 1.37, m | 25.2, t | 1.34, m |
| 1.45, m | 1.44, m | |||
| 16 | 31.7, t | 1.31, m | 31.7, t | 1.29, m |
| 17 | 22.6, t | 1.32, m | 22.6, t | 1.30, m |
| 18 | 14.1, q | 0.91, t (8.0) | 14.0, q | 0.89, t (8.0) |
| 1' | 162.8, s | 164.3, s | ||
| 2' | 76.5, s | 73.6, s | ||
| 3' | 35.6, t | 4.32, br d (18.0) | 39.9, t | 3.74, d (17.0) |
| 3.25 d (18.0) | 3.44, d (17.0) | |||
| 4' | 128.1, s | 131.4, s | ||
| 5' | 125.3, d | 7.33, d (7.0) | 116.2, d | 6.82, d (7.0) |
| 6' | 125.9, d | 7.21, t (7.0) | 129.3, d | 7.17, t (7.0) |
| 7' | 128.8, d | 7.34, t (7.0) | 118.4, d | 6.92, d (7.0) |
| 8' | 115.8, s | 7.93, d (7.0) | 146.4, s | |
| 9' | 138.1, s | 126.7, s | ||
| 2-SMe | 15.1, q | 2.36, s | ||
| 2'-SMe | 14.2, q | 2.20, s | ||
| 8'-OH | 10.03, s | |||
| 13-OH | NDc | NDc | ||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download