Abstract
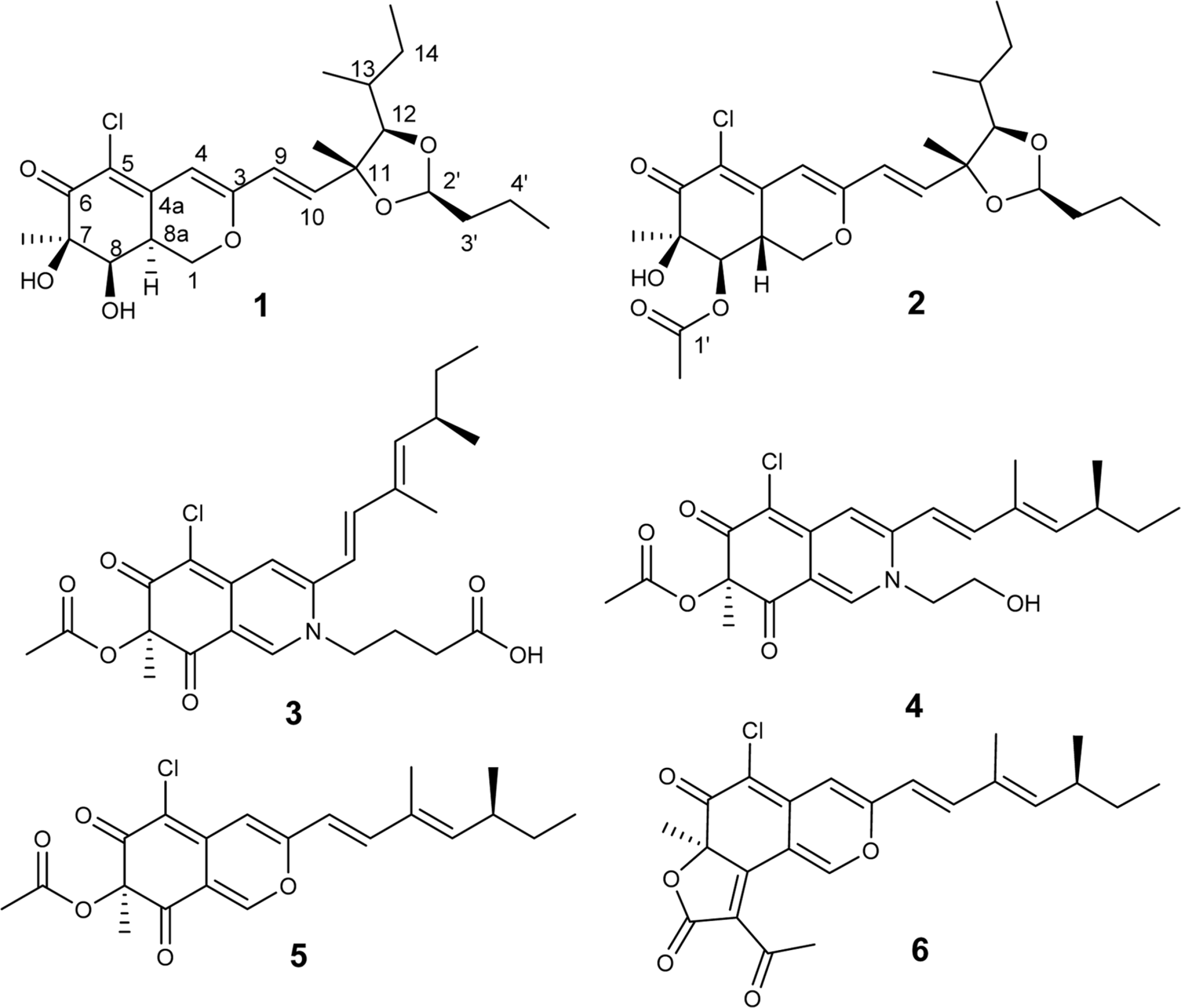
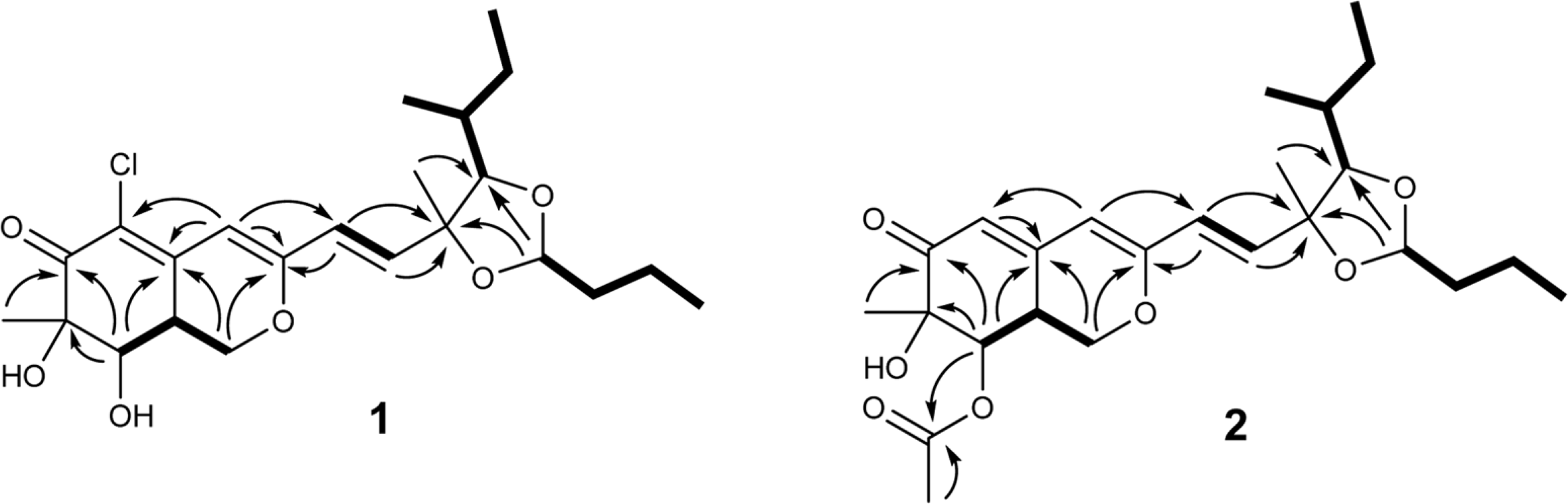
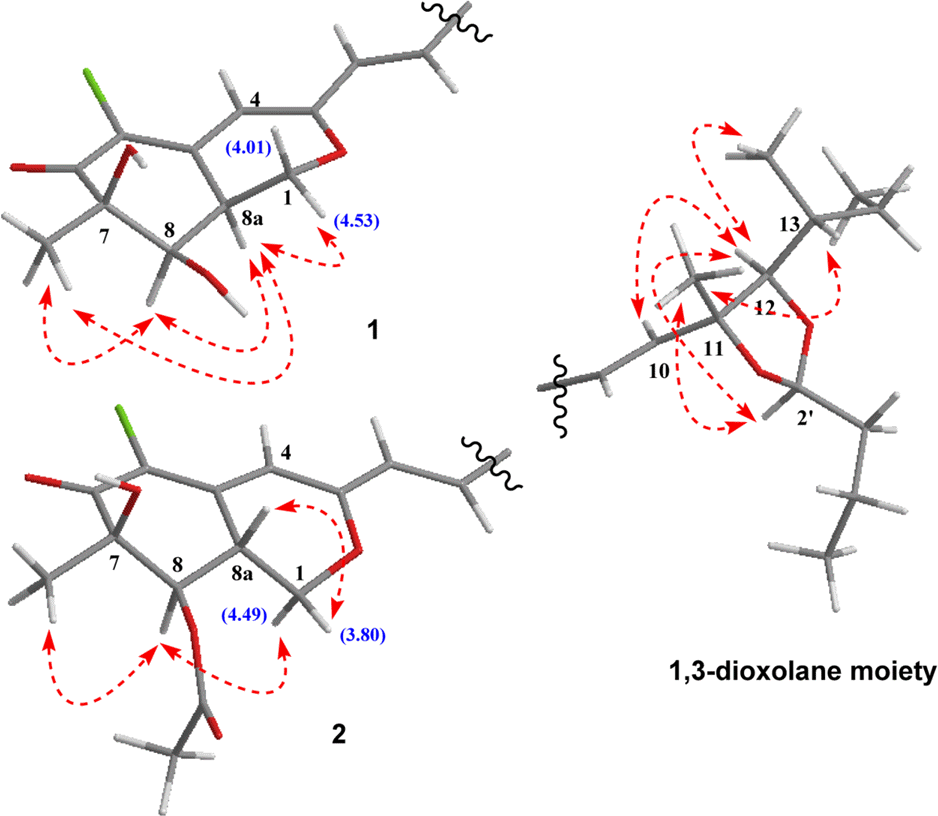
Two new azaphilone derivatives containing 1,3-dioxolane moiety, penidioxolanes A (1) and B (2), were isolated from marine-derived fungus Penicillium sp. KCB12C078, together with four known compounds (3-6) by chemical investigation. Compounds 1 - 6 were isolated by combination of silica gel, ODS column chromatography and preparative HPLC. Their structures were determined by analysis of spectroscopic data including 1D-, 2D-NMR, and MS techniques. The isolates were evaluated against cancer cell growth inhibition effects and antimicrobial activity.
Go to : 
REFERENCES
(1). Bérdy J. J.Antibiot. 2005; 58:1–26.
(2). Fenical W., Jensen P. R.Nat. Chem. Biol. 2006; 2:666–673.
(3). Faulkner D. J.Nat. Prod. Rep. 1998; 15:113–158.
(4). Satpute S. K., Banat I. M., Dhakephalkar P. K., Banpurkar A. G., Chopade B. A.Biotechnol. Adv. 2010; 28:436–450.
(5). Osmanova N., Schultze W., Ayoub N.Phytochem. Rev. 2010; 9:315–342.
(6). Gao J. M., Yang S. X., Qin J. C.Chem. Rev. 2013; 113:4755–4811.
(7). Quang D. N., Hashimoto T., Fournier J., Stadler M., Radulovi cí N., Asakawa Y.Tetrahedron. 2005; 61:1743–1748.
(8). Kanokmedhakul S., Kanokmedhakul K., Nasomjai P., Louangsy-souphanh S., Soytong K., Isobe M., Kongsaeree P., Prabpai S., Suksamrarn A. J.Nat. Prod. 2006; 69:891–895.
(9). Yu B. Z., Zhang G. H., Du Z. Z., Zheng Y. T., Xu J. C., Luo X. D.Phytochemistry. 2008; 69:2523–2526.
(10). Quang D. N., Hashimoto T., Tanaka M., Stadler M., Asakawa Y.Phytochemistry. 2004; 65:469–473.
(11). Li J. J., Shang X. Y., Li L. L., Liu M. T., Zheng J. Q., Jin Z. L.Molecules. 2010; 15:1958–1966.
(12). Dong J., Zhou Y., Li R., Zhou W., Li L., Zhu Y., Huang R., Zhang K.FEMS. Microbiol. Lett. 2006; 264:65–69.
(13). Yasukawa K., Takahashi M., Natori S., Kawai K., Yamazaki M., Takeuchi M., Takido M.Oncology. 1994; 51:108–112.
(14). Michael A. P., Grace E. J., Kotiw M., Barrow R. A.Aust. J. Chem. 2003; 56:13–15.
(15). Arai N., Shiomi K., Tomoda H., Tabata N., Yang D. J., Masuma R., Kawakubo T., Omura S. J.Antibiot. 1995; 48:696–702.
(16). Curtin T. P., Reilly J.Biochem. J. 1940; 34:1419–1421.
(17). Holker J. S. E., Ross W. J., Staunton J., Whalley W. B. J.Chem. Soc. 1962; 4150–4154.
(18). Gray R. W., Whalley W. B.Chem. Commun. 1970; 12:762.
(19). Yang D. J., Tomoda H., Tabata N., Masuma R., Omura S. J.Antibiot. 1996; 49:223–229.
(20). Matsuzaki K., Tanaka H., Omura S. J.Antibiot. 1995; 48:708–713.
(21). Arunpanichlert J., Rukachaisirikul V., Sukpondma Y., Phongpaichit S., Tewtrakul S., Rungjindamai N., Sakayaroj J.Chem. Pharm. Bull. 2010; 58:1033–1036.
Go to : 
Table 1.
1H (900 MHz) and 13C (225 MHz) NMR data of 1 and 2 in DMSO-d6




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download