Abstract
Purpose:
The prevalence rate of the BRAF mutation in papillary thyroid cancer (PTC) is as high as about 52 to 83% in Korea. Preoperative detection of BRAF mutation on fine needle aspiration cytology (FNAC) slides may help the surgeon make better therapeutic decisions. The present study aims to assess the feasibility of the mutant allele specific amplification (MASA) and restriction fragment length polymorphism (RFLP) method with using conventional FNAC slides and we also wanted to evaluate the clinical role of preoperatively detecting BRAF mutation.
Methods:
We extracted the genomic DNA from 59 FNAC slides and performed direct sequencing (DS) for detecting BRAF mutation. We could use only 17 slides for the MASA method and 6 slides for the RFLP method due to the shortage of extracted DNA. Additionally, we retrospectively analyzed the cases for which a histological diagnosis could be made.
Results:
Genomic DNA was extracted from 23 out of the 59 FNAC slides. The BRAF mutation status could be assessed via DS in 33 out of the 59 FNAC slides. The concordance between the MASA method and DS and the RFLP method and DS was 36.3% and 66.7% respectively. The positive and negative predictive value of the 13 indeterminate nodules was 87.5% and 20%, respectively. We could not find any association between the BRAF mutations and the alleged risk factors of PTC.
Conclusion:
We believe that the purity and the amount of the DNA template must be increased to detect BRAF mutation with using a FNAC slide. Preoperative detection of the BRAF mutation on a FNAC slide may refine the cytological diagnosis, but the application of assessing BRAF mutation as a prognostic marker is debatable.
Go to : 
REFERENCES
1.Davies H., Bignell GR., Cox C., Stephens P., Edkings S., Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002. 417:949–54.

3.Garnett MJ., Marais R. Guilty as charged: B-RAF is a human oncogene Cancer Cell. 2004. 6:313–9.
4.Cohen Y., Xing M., Mambo E., Guo Z., Wu G., Trink B, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003. 95:625–7.

5.Soares P., Trovisco V., Rocha AS., Lima J., Castro P., Preto A, et al. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003. 22:4578–80.

6.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007. 28:742–62.

7.Shin HR., Jung KW., Won YJ., Park JG. 2002 annual report of the Korea Central Cancer Registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004. 36:103–14.

8.Kim TY., Kim WB., Song JY., Rhee YS., Gong G., Cho YM, et al. The BRAF mutation is not associated with poor prognostic factors in Korean patients with conventional papillary thyroid microcarcinoma. Clin Endocrinol (Oxf). 2005. 63:588–93.
9.Kim KH., Kang DW., Kim SH., Seong IO., Kang DY. Mutations of the BRAF gene in papillary thyroid carcinoma in a Korean population. Yonsei Med J. 2004. 45:818–21.

10.Chow LS., Gharib H., Goellner JR., van Heerden JA. Nondiagnostic thyroid fine-needle aspiration cytology: management dilemmas. Thyroid. 2001. 11:1147–51.

11.Usami S., Abe S., Shinkawa H., Inoue Y., Yamaguchi T. Rapic mass screening method and counseling for the 1555A→G mitochondrial mutation. J Hum Genet. 1999. 44:304–7.
12.Chung YS., Choe JH., Lee KE., Park WS., Kim HY., Han W, et al. Detection of BRAF mutation on fine needle aspiration biopsy specimens using the colorimetric mutation detection method. J Korean Surg Soc. 2008. 74:25–33.
13.Sommer SS., Cassady JD., Sobell JL., Bottema CDK. A novel method for detecting point mutations or polymorphisms and its application to population screening for carriers of phenylketonuria. Mayo Clin Proc. 1989. 64:1361–72.

14.Hayashi N., Arakawa H., Nagase H., Yanagisawa A., Kato Y., Ohta H, et al. Genetic diagnosis identifies occult lymph node metastases undetectable by the histopathological method. Cancer Res. 1994. 54:3853–6.
15.Sapio MR., Posca D., Troncone G., Pettinato G., Palombini L., Rossi G, et al. Detection of BRAF mutation in thyroid papillary carcinomas by mutant allele-specific PCR amplification (MASA). Eur J Endocrinol. 2006. 154:341–8.

16.Kumagai A., Namba H., Akanov Z., Saenko VA., Meirmanov S., Ohtsuru A, et al. Clinical implications of pre-operative rapid BRAF analysis for papillary thyroid cancer. Endocr J. 2007. 54:399–405.

17.Chung KW., Yang SK., Lee GK., Kim EY., Kwon S., Lee SH, et al. Detection of BRAFV600E mutation on fine needle aspiration specimens of thyroid nodule refines cyto-pathology diagnosis, especially in BRAF600E mutation-prevalent area. Clin Endocrinol (Oxf). 2006. 65:660–6.
19.Oertel YC., Burman K., Boyle L., Ringel M., Wartofsky L., Shmookler B, et al. Integrating fine-needle aspiration into a daily practice involving thyroid disorders: the Washington Hospital Center approach. Diagn Cytopathol. 2002. 27:120–2.

20.Lee JH., Lee ES., Kim YS. Clinicopathologic significance of BRAF V600E mutation in papillary carcinomas of the thyroid: a meta-analysis. Cancer. 2007. 110:38–46.
21.Lupi C., Giannini R., Ugolini C., Proietti A., Berti P., Minuto M, et al. Association of BRAF V600E mutation with poor clinicopathological outcomes in 500 consecutive cases of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2007. 92:4085–90.

22.Xing M., Westra WH., Tufano RP., Cohen Y., Rosenbaum E., Rhoden KJ, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab. 2005. 90:6373–9.

23.Kim TY., Kim WB., Rhee YS., Song JY., Kim JM., Gong G, et al. The BRAF mutation is useful for prediction of clinical recurrence in low-risk patients with conventional papillary thyroid carcinoma. Clin Endocrinol (Oxf). 2006. 65:364–8.

24.Kebebew E., Weng J., Bauer J., Ranvier G., Clark OH., Duh QY, et al. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann Surg. 2007. 246:466–71.

25.Park SY., Park YJ., Lee YJ., Lee HS., Choi SH., Choe G, et al. Analysis of differential BRAF(V600E) mutational status in multifocal papillary thyroid carcinoma: evidence of independent clonal origin in distinct tumor foci. Cancer. 2006. 107:1831–8.
Go to : 
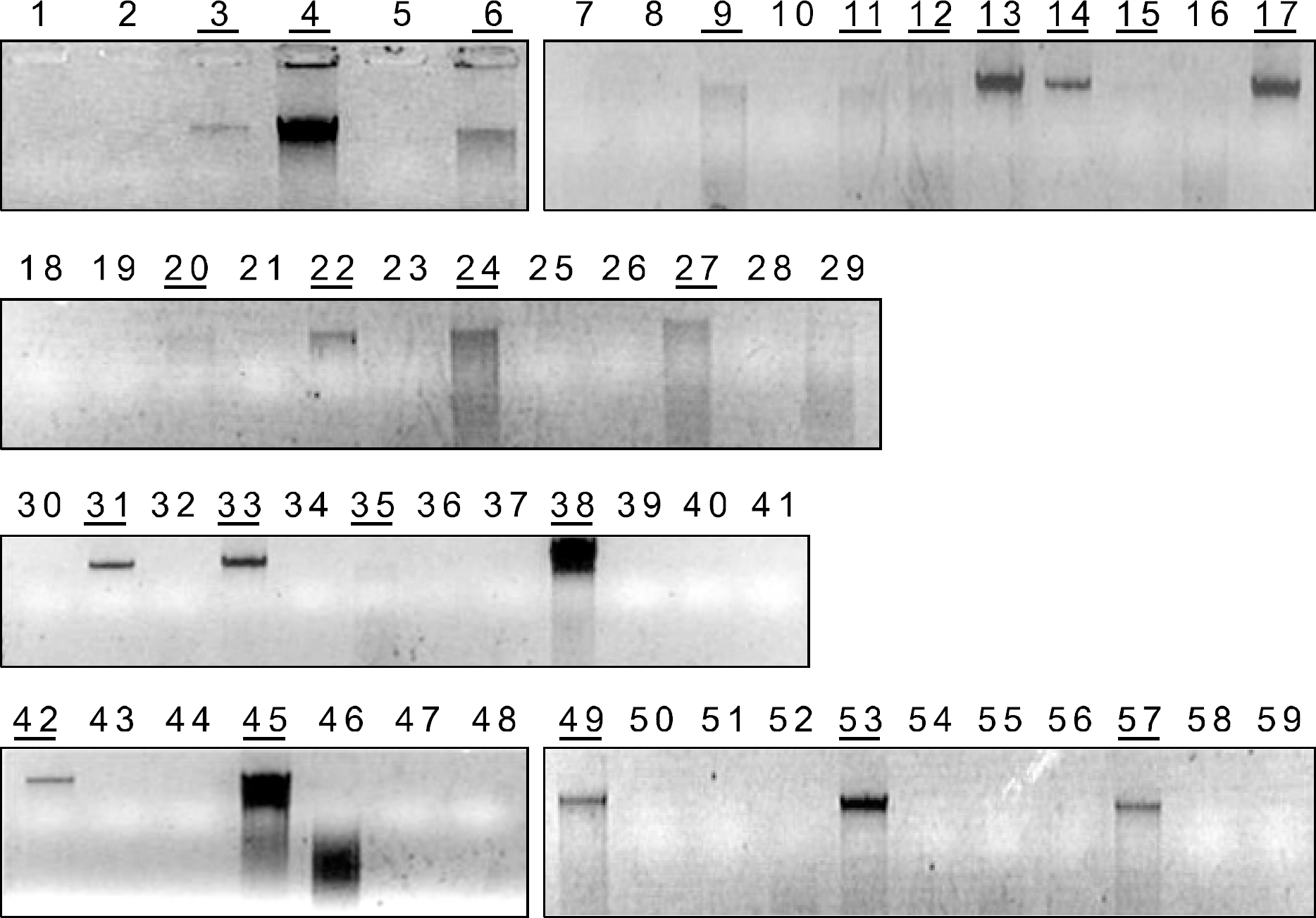 | Fig. 2BRAF exon15 PCR amplification. Which samples have enough and pure DNA to be sequenced might not be determined on electrophoresis gel. |
 | Fig. 3MASA products showing low sensitivity for BRAF mutation. Faint bands on No. 1, 3, 16 and 17 indicate the presence of BRAF mutation (Underlined numbers of samples have been proved for positive BRAF mutation by direct sequencing). |
Table 1.
Results of direct sequencing with fine needle aspiration cytology slides
| No.∗ | Direct sequencing | No. | Direct sequencing | No. | Direct sequencing |
|---|---|---|---|---|---|
| 1 | Mutant type | 21 | Mutant type | 41 | X† |
| 2 | Mutant type | 22 | Wild | 42 | Wild |
| 3 | Mutant type | 23 | X | 43 | X |
| 4 | Mutant type | 24 | Mutant type | 44 | X |
| 5 | X† | 25 | X | 45 | Mutant type |
| 6 | Mutant type | 26 | Wild | 46 | X |
| 7 | Mutant type | 27 | Mutant type | 47 | X |
| 8 | X | 28 | Mutant type | 48 | X |
| 9 | Mutant type | 29 | Mutant type | 49 | Mutant type |
| 10 | Mutant type | 30 | X | 50 | X |
| 11 | Mutant type | 31 | Mutant type | 51 | X |
| 12 | X | 32 | Wild | 52 | X |
| 13 | Mutant type | 33 | Wild | 53 | X |
| 14 | Wild | 34 | Wild | 54 | Mutant type |
| 15 | Wild | 35 | Wild | 55 | X |
| 16 | Mutant type | 36 | X | 56 | X |
| 17 | Wild | 37 | Mutant type | 57 | Mutant type |
| 18 | X | 38 | Mutant type | 58 | X |
| 19 | X | 39 | X | 59 | X |
| 20 | X | 40 | X |
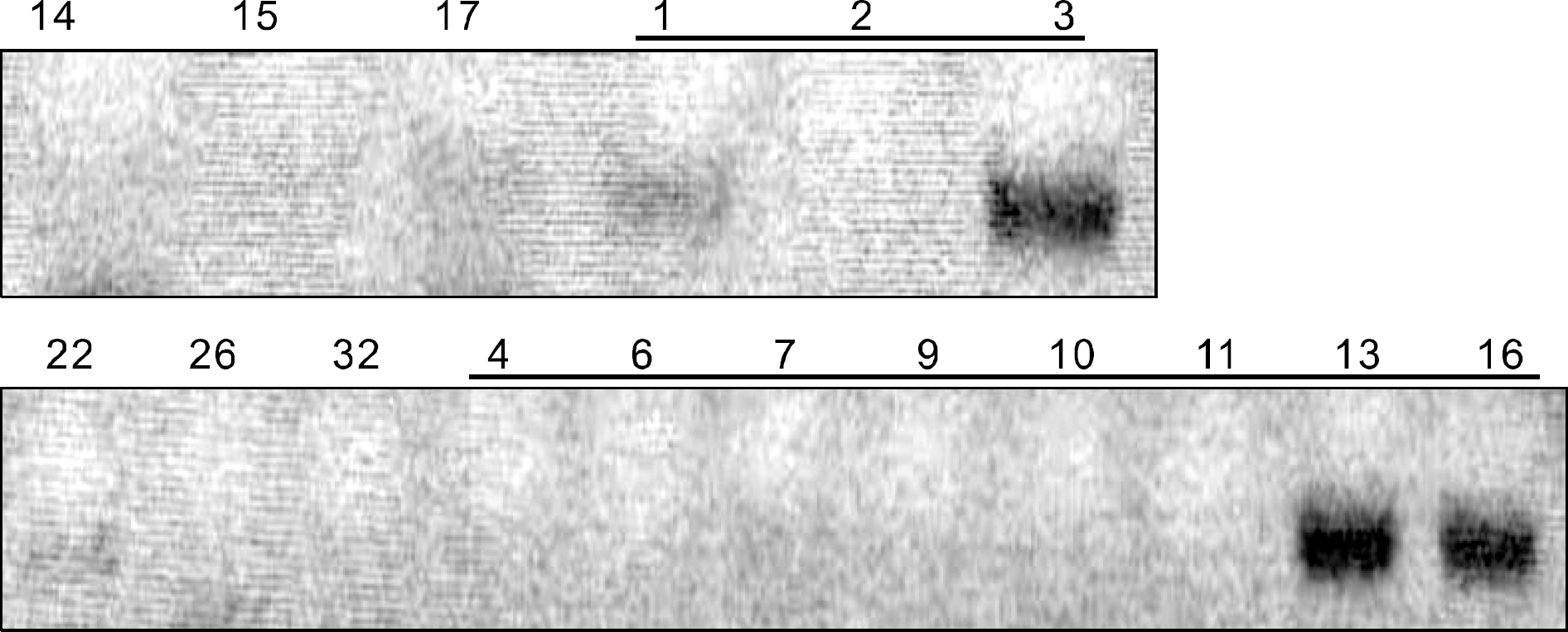 | Fig. 4RFLP products showing insecure results for BRAF mutation (Underlined numbers of samples have been proved for positive BRAF mutation by direct sequencing). |
Table 2.
Association between cytological diagnosis and BRAF mutation status
| Cytological diagnosis | BRAF status∗ | |||
|---|---|---|---|---|
| Mutant type | Wild type | No information | Total | |
| Benign | 1† | 2 | 4 | 7 |
| Suspicious for FN‡ | 1§ | 2 | 5 | 8 |
| Suspicious for PTC∥ | 8 | 3 | 4 | 15 |
| Malignant (PTC) | 13 | 3 | 9 | 25 |
| Inadequate | 0 | 0 | 4 | 4 |
| Total | 23 | 10 | 26 | 59 |
Table 3.
Results of the cytological diagnosis and the number of patients who received surgery in Seoul National University Hospital
| Cytological diagnosis | Number of patients received surgery (n=46) | Number of lost patients (n=11) |
|---|---|---|
| Benign | 3 | 3 |
| Suspicious for FN∗ | 8 | 0 |
| Suspicious for PTC† | 12 | 3 |
| Malignant (PTC) | 20 | 4 |
| Inadequate | 3 | 1 |
Table 4.
BRAF mutation status of fine needle aspiration cytology slides with malignant and indeterminate result on cytology and postoperative histological diagnosis
| Cytological diagnosis | BRAF status | Histological diagnosis (Number) |
|---|---|---|
| Malignant | Mutant | PTC∗ (11) |
| Wild | PTC (2) | |
| Suspicious PTC | Mutant | PTC (7) |
| Wild | PTC (2), Minimal invasive FTC† (1) | |
| Suspicious FN‡ | Mutant | Nodular hyperplasia (1) |
| Wild | Minimal invasive FTC (1), Nodular hyperplasia (1) |
Table 5.
Association between BRAF mutation status and clinico pathological characteristics in 22 papillary thyroi cancer patients
| BRAF mutation (+) | BRAF mutation (−) | P value | |
|---|---|---|---|
| Number | 18 | 4 | |
| Female:Male | 14:4 | 14:1 | 0.905∗ |
| Age (range) | 51.3 (34∼74) | 43.5 (39∼49) | 0.233† |
| Tumor size (cm) | 1.33±0.76 | 1.05±0.7 | 0.494† |
| LN metastasis | 6 (33.3%) | 2 (50%) | 0.602∗ |
| Perithyroidal extension | 14 (77.8%) | 3 (75%) | 1.000∗ |
| Multiplicity | 7 (38.9%) | 3 (75%) | 0.293∗ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download