Abstract
Purpose
Today, many urologists use nucleic acid amplification tests (NAAT) in diagnosis of Chlamydia trachomatis infection in Korea. A new variant of C. trachomatis with a deletion in the cryptic plasmid, which cannot be detected using commercial tests targeting the deleted DNA sequences, has been found in Sweden. Therefore, the partial deletion of cryptic plasmid DNA means that the diagnostic standards cannot detect chlamydial infection any more in cases of new mutants. The mutant type has been prevalent in Sweden, however, its incidence was not high in other countries such as France, Holland, and Denmark. In or to study the existence of this mutant C. trachomatis in Korea, we developed new primer sets for detection of this mutation.
Materials and Methods
We collected the first voided urine from male urethritis patient from April 2012 to August 2013 (Dankook University Hospital, Cheonan, Korea). We used the 25 confirmed C. trachomatis-positive specimens by using KL1 and KL2 primers for C. trachomatis and tested the existence of mutant chlamydial infection with the newly developed primer sets.
REFERENCES
1. Groseclose SL, Zaidi AA, DeLisle SJ, Levine WC, St Louis ME. Estimated incidence and prevalence of genital Chlamydia trachomatis infections in the United States, 1996. Sex Transm Dis. 1999; 26:339–44.

2. Stamm WE. Chlamydia trachomatis infections of the adult. Holmes KK, Sparling PF, Mardh P, Lemon SM, Stamm WE, Piot P, editors. Sexually transmitted diseases. 3rd ed.New York: McGraw-Hill;1999. p. 407–22.
3. Lee G, Sohng I. Cryptic plasmid amplification of Chlamydia trachomatis at a Korean Health Center for Female Commercial Sex Workers. Korean J Urol. 2006; 47:37–41.
4. Unemo M, Olcén P, Agné-Stadling I, Feldt A, Jurstrand M, Herrmann B, et al. Experiences with the new genetic variant of Chlamydia trachomatis in Orebro county, Sweden - proportion, characteristics and effective diagnostic solution in an emergent situation. Euro Surveill. 2007; 12:E5–6.
5. Jurstrand M, Christerson L, Klint M, Fredlund H, Unemo M, Herrmann B. Characterisation of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex Transm Infect. 2010; 86:56–60.

6. Reinton N, Moi H, Bjerner J, Moghaddam A. The Swedish Chlamydia mutant nvC trachomatis in Norway. Tidsskr Nor Laegeforen. 2010; 130:380–1.
8. de Barbeyrac B, Raherison S, Cado S, Normandin F, Clerc M, Clairet V, et al. French situation concerning the Swedish Chlamydia trachomatis variant. Euro Surveill. 2007; 12:E11–2.

9. Morré SA, Catsburg A, de Boer M, Spaargaren J, de Vries HJ, Schirm J, et al. Monitoring the potential introduction of the Swedish Chlamydia trachomatis variant (swCT) in the Netherlands. Euro Surveill. 2007; 12:E9–10.

10. National guideline for the management of Chlamydia trachomatis genital tract infection. Clinical Effectiveness Group (Association of Genitourinary Medicine and the Medical Society for the Study of Venereal Diseases). Sex Transm Infect. 1999; 75(Suppl 1):S4–8.
11. Pasternack R, Vuorinen P, Pitkäjärvi T, Koskela M, Miettinen A. Comparison of manual Amplicor PCR, Cobas Amplicor PCR, and LCx assays for detection of Chlamydia trachomatis infection in women by using urine specimens. J Clin Microbiol. 1997; 35:402–5.

12. Davis JD, Riley PK, Peters CW, Rand KH. A comparison of ligase chain reaction to polymerase chain reaction in the detection of Chlamydia trachomatis endocervical infections. Infect Dis Obstet Gynecol. 1998; 6:57–60.
13. Lee G, Park J, Kim B, Kim SA, Yoo CK, Seong WK. OmpA genotyping of Chlamydia trachomatis from Korean female sex workers. J Infect. 2006; 52:451–4.

14. Byun Y, Park HY, Kim H, Lee G. Differences in Chlamydia trachomatis cryptic plasmid loads in two types of female commercial sex workers in Korea. Jpn J Infect Dis. 2008; 61:146–7.
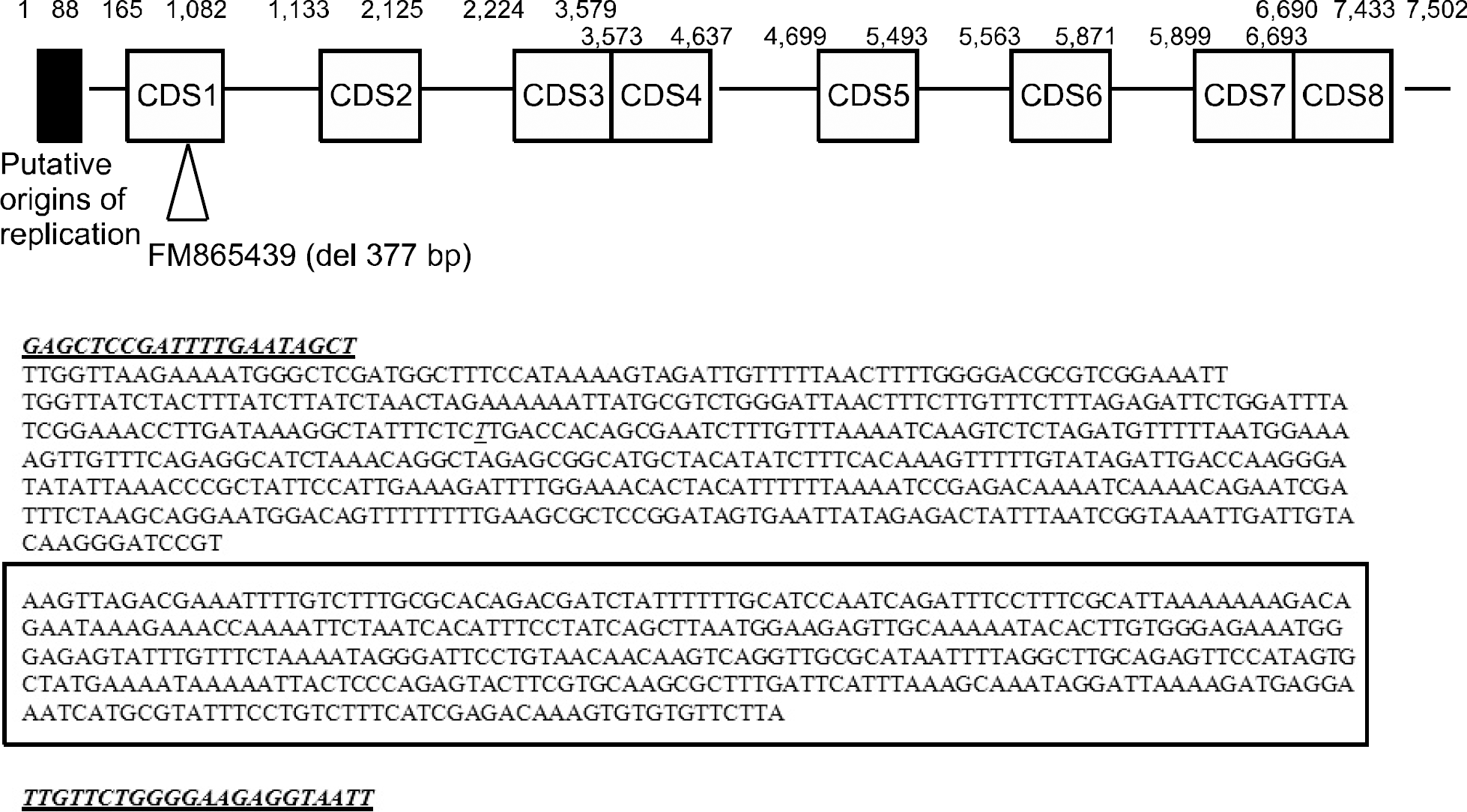
Fig. 1.
Schematic view of the partial Chlamydia trachomatis cryptic plasmid DNA sequences (reference sequence: HE603233.1). Chlamydia trachomatis plasmid pSW2, strain Sweden2, serovar E (reference sequence; FM8654) revealed 1 377-bp deletion in CDS1 region (box). CDS; coding sequence.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download