Abstract
Background
Mitochondria are major cellular sources of reactive oxygen species (ROS) generation which can induce mitochondrial DNA damage and lead to carcinogenesis. The mitochondrial 10398A>G alteration in NADH-dehydrogenase subunit 3 (ND3) can severely impair complex I, a key component of ROS production in the mitochondrial electron transport chain. Alteration in ND3 10398A>G has been reported to be linked with diverse neurodegenerative disorders and cancers. The aim of this study was to find out the association of mitochondrial ND3 10398A>G alteration in brain tumor of Malaysian patients.
Methods
Brain tumor tissues and corresponding blood specimens were obtained from 45 patients. The ND3 10398A>G alteration at target codon 114 was detected using the PCR-RFLP analysis and later was confirmed by DNA sequencing.
Results
Twenty-six (57.8%) patients showed ND3 10398A>G mutation in their tumor specimens, in which 26.9% of these mutations were heterozygous mutations. ND3 10398A>G mutation was not significantly correlated with age, gender, and histological tumor grade, however was found more frequently in intra-axial than in extra-axial tumors (62.5% vs. 46.2%, p<0.01).
Conclusion
For the first time, we have been able to describe the occurrence of ND3 10398A>G mutations in a Malaysian brain tumor population. It can be concluded that mitochondrial ND3 10398A>G alteration is frequently present in brain tumors among Malaysian population and it shows an impact on the intra-axial tumors.
Brain tumor is a leading cause of solid tumor cancer death worldwide among children and adolescents (aged birth to 19 years), approximately accounting 4,630 new cases each year. The worldwide review report of brain tumor by de Robles and colleagues [1] using a random-effects model determined that the overall incidence rate of all brain tumors to be 10.82 (95% CI: 8.63–13.56) per 100,000 person-years. The National Brain Tumor Society in United State estimated that 78,980 individuals (55,150 benign and 23,830 malignant) would be diagnosed with primary brain tumors in 2018 and that 16,616 of those individuals would die due to this tumor [2]. In Malaysia, according to the regional cancer registers, it was estimated that the age-standardised incidence rate of brain and central nervous system (CNS) tumors in 2012 to be 2.8 per 100,000 population/year with cumulative rate 0.3%. This rate was increasing faster until it became the third commonest pediatric cancer in Malaysia after leukemia and lymphoma [34].
Mitochondria appear to play a number of important functions within the cell as they generate most of the adenosine triphosphate (ATP) via the oxidative phosphorylation system (OXPHOS), produce free radicals through dysfunction of the electron transport chain, and regulate cell death [56]. Imperfections of mitochondrial function have long been recognized to have an impact on the development and progression of cancer [7]. Cancer cells exhibit genetic instability, with alterations such as mutation, deletions, insertions and microsatellite instability in both nuclear and mitochondrial genome. So far, mitochondrial DNA (mtDNA) researches in development of cancer have intensively been focused due to its 10 times more sensitive to mutations in comparison to nuclear DNA, inefficient repair mechanisms and close proximity to the reactive oxygen species (ROS)-damaging electron transport system [891011].
The human mtDNA encodes for 13 polypeptides that are part of four of the five respiratory chain (RC) complexes in the OXPHOS system: ND1-6, ND4L (complex I), Cyt b (complex III), CO1-3 (complex IV), ATP6 and 8 (complex V) and also 22 tRNAs and 2 rRNAs which are specific for mitochondrial translation. Mutations in the RC complexes genes have impact on the function of the OXPHOS complexes, and may cause OXPHOS system deficiencies [12] and these mitochondrial RC defects have been reported in human solid tumors [13]. Up to now, various type of cancer have been reported to be associated with mtDNA mutations in all of the 13 protein coding genes, in both the 12S and 16S rRNA genes, in each of the 22 tRNA genes, and in the non-coding displacement-loop (D-loop) [814151617181920].
The 10398 nucleotide position in the human mtDNA NADH dehydrogenase subunit 3 (ND3) of complex I of the electron transport chain is highly polymorphic. This polymorphism (A to G transition) turns the amino acid substitution from threonine into alanine. It has been suggested that altered 10398A>G may lead to a decreased efficiency of the complex I function and subsequently enhanced cellular ROS production [21222324]. This condition may result in increased oxidative stress which will further accumulate more damage to mtDNA that can initiate and promote the tumorigenesis process. There have been reports on the presence of 10398A>G polymorphism in neurodegenerative disorders such as Alzheimer's [2526] and Parkinson's disease [262728]. The involvement of 10398A>G mutation as a risk factor in breast cancer has also been previously studied but the findings have been controversial [2122293031].
DNA alterations in mitochondria are believed to become fast hotspots of cancer research. In our previous studies, we discovered that brain tumor tissues had a high frequency (51% were identified in 49 patients) of somatic mutations in the non-coding D-loop region of mtDNA [32]. Novel mutations that have not been reported previously at D-loop region such as 176 A-deletion, 476C>A, 566C>A, and 16405 A-deletion were also identified in our patients. These findings lead us to further the study to seek if the mutation occurs in the other regions of mtDNA. In the present study, we tried to focus on the mtDNA coding region specifically ND3 10398A>G mutation in brain tumor, since there are no published data available on that tumor at this moment. Therefore, the aim of the present study was to examine the ND3 10398A>G alteration status in brain tumor of Malaysian patients and determine its association with clinicopathological features.
The present study was approved by Research Ethics Committees of Neuroscience Department, Universiti Sains Malaysia (FWA Reg. No: 00007718; IRB Reg. No: 00004494). A total of 45 Malay brain tumor patients who had undergone neurosurgical procedures were enrolled into this study. Patients who had received previous radiotherapy to the brain or chemotherapy for any reason were excluded. The histopathological diagnosis of brain tumors was confirmed by a consultant neuropathologist and examined according to World Health Organization (WHO) criteria [33]. All fresh tumor tissues and their corresponding blood specimens (peripheral blood from the same patients as the control) were obtained with the informed consent of the patients and they were stored frozen at −80℃ until analysis. For comparative purposes, 20 archival paraffin-embedded tissues that confirmed normal human brain tissues obtained from autopsies of accident victims were used in this study. Besides that, blood samples from 40 unrelated healthy controls free of brain tumors or other major of CNS tumors were also collected, as control.
All total DNA extraction was carried out using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) by following the manufacturer's recommended protocol. The concentration and quality of extracted DNA was measured using the NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and 1% agarose gel electrophoresis (Thermo Scientific Inc., San Jose, CA, USA).
A region of mtDNA spanning 10147–10569 encoding for ND3, was PCR-amplified using the specific primer pair: forward, 5′-ACATAGAAAAATCCACCCCT-3′ and reverse, 5′-CTAGGCATAGTAGGGAGGAT-3′. The expected amplicon length is 423 bp. The reaction mixture (50 µL) in the PCR contained: 100 ng DNA template, 200 µM of each dNTP, 20 pmol of each primer, 10 µL of 5 X Phusion HF buffer (Thermo Scientific, Inc.) and 2 U Phusion high-fidelity DNA polymerase (Thermo Scientific, Inc.). The PCR cycling conditions were: 98℃ for 30s, 50℃ for 30s, and 72℃ for 40s, for 35 cycles in a SureCycler 8800 Thermal Cycler (Agilent Technologies, Santa Clara, CA, USA). There was a 3 min pre-incubation at 98℃ before starting the cycles, and 5 min at 72℃ after the completion of the cycles.
The 423bp products of ND3 obtained were digested with FastDigest DdeI (HpyF3I) restriction endonuclease (Thermo Scientific Inc.) according to manufacturer's instructions. Digestions were carried out in a total volume of 15 µL. The reaction mixture consisted of 3 µL of PCR product, 1 µL of restriction buffer (10X), 0.5 µL of restriction enzyme (10 U/µL) and volume adjusted with sterile distilled water. Then the digestion reaction was incubated for 20 min at 37℃. The restriction fragments were analysed on 2% agarose gel, stained with ethidium bromide and visualized under ultraviolet light.
Direct sequencing was used to validate the genotyping results of ND3 and PCR-restriction fragment length polymorphism (RFLP) assay. Purified PCR products were sequenced using the same primers as that described in the PCR amplification. Sequencing was performed using a Big Dye Terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, CA, USA) on an ABI Prism 3700 DNA Analyser automated sequencer (Applied Biosystems Inc.).
The statistical analysis was performed using GraphPad Prism software version 7 (Graphpad Software, San Diego, CA, USA). The Fisher's exact test was used to calculate the significance of relationship between ND3 10398A>G mutation and clinicopathological parameters of brain tumor. Differences were considered statistically significant at a p value of <0.05.
Clinicopathologic features including the age, gender, and histological tumor types are provided in Table 1. To find out the association of mitochondrial 10398A>G mutation in ND3 in brain tumor, 45 specimens of brain tumors were collected from Malaysian patients. The age of patients at the time of diagnosis ranged from 3 to 68 years, with an average age of 41.9±18.3 years. There were 26 (57.8%) males and 19 (42.2%) females. According to histopathologic type, greatest number of cases was in glioblastoma multiforme (IV) (37.8%), followed by meningioma (I) (24.4%), anaplastic oligodendroglioma (III) (8.9%), anaplastic ependymoma (III) (6.7%), pilocytic astrocytoma (I) (4.4%), astrocytoma (II) (4.4%), ependymoma (II) (4.4%), schwannoma (I) (4.4%), and medulloblastoma (IV) (4.4%). Furthermore, all these brain tumors were graded into two categories, extra-axial (28.9%) and intra-axial (71.1%) tumor groups, based on the WHO pathological findings.
A region of mtDNA encoding for ND3, spanning the 10398A>G mutation hotspot of ND3, was successfully amplified by PCR using the specific primers. A single PCR amplicon, corresponding in size to the predicted 422-bp fragment was observed (Fig. 1).
The experiment was designed to use the HpyF3I (DdeI) restriction enzyme, which digested the ND3 PCR amplicons encompassing 10398A>G in different fragments. The RFLP profile obtained three fragments of 213-bp, 130-bp, and 80-bp for the wild-type alleles. Digestion of an A10398G mutant sample results in four bands of 175-bp, 130-bp, 80-bp, and 38-bp. Besides that, digestion of a heterozygous sample results in five bands: (three bands: a 213-bp band from the normal allele and a band with the 175-bp and 38-bp fragments from the mutant; two bands: 130-bp and 80-bp) (Fig. 2).
To verify the RFLP results, PCR products were sequenced. DNA sequencing was consistent with the pattern obtained by RFLP assay in all cases (Fig. 3). This ND3 mutation A to G transition at nucleotide position 10398, converts the amino acid change from threonine codon (ACC) into an alanine codon (GCC) at amino acid position 114 (T114A).
This study revealed that 57.8% (26 out of 45) of the patients carried ND3 mutation at 10398A>G (Fig. 4A). Moreover, 26.9% (n=7/26) of these mutations were heterozygous mutations (Fig. 4B). None of the patient-matched blood samples and all control groups (including 20 autopsy brain samples of healthy subjects dead by accidents) showed ND3 10398A>G mutation.
The correlation between ND3 10398A>G mutation status with gender, age, and histological tumor types/clinical grade of the patient was described in Table 2. Regarding the tumor grading, brain tumors were divide into two classifications, extra-axial and intra-axial tumors groups, by their location. No statistically significant correlation was found between both tumors groups and ND3 10398A>G in this study. ND3 10398A>G mutation was detected significantly more frequently (p<0.01) in intra-axial (n=20/32, 62.5%) than extraaxial tumors (n=6/13, 46.2%).
In both groups ≤40 and >40 years, the correlation was determined to be non-significant (p=0.212) although ND3 10398A>G mutations were found to be more evident in the age group >40 years (n=19/29, 65.5%) than the age group ≤40 (n=7/16, 43.8%).
There was no significant association between the ND3 10398A>G mutation status and the gender (p=0.360). In this study, male patients (n=17/26, 65.4%) exhibited a high rate of ND3 10398A>G mutation than female patients (n=9/19, 47.4%).
Alterations in mtDNA which may lead to OXPHOS system destabilization seem to be particularly crucial because 13 proteins encoded by mtDNA are essential for complex I, and III–V RC assembly and these enzymatic complexes defects have been reported in various human diseases [35]. Abnormalities in mtDNA have proven to be associated with diabetes mellitus, Parkinson's disease, Alzheimer's disease, epilepsy, sensorineural deafness, and a variety of syndromes involving muscles and the CNS [3637]. Moreover, 25–80% of somatic mutations in mtDNA are discovered in a variety of human cancers [38].
In the present study, mutational screening for mitochondrial 10398A>G alteration in ND3 was performed in the Malaysian brain tumor patients, as far as we know, has not been investigated. To the best of our knowledge, the initial finding of 10398A>G change was described as a missense mutation by Kosel's group in Parkinson's disease [27]. Several studies described the association of 10398A>G mutation with human breast cancer risk but their outcomes were considered controversial. For the past few years, scientists in breast cancer research have paid intensively attention to the 10396A>G mutation as a plausible candidate marker for increased breast cancer susceptibility [212939404142]. In a study that involved African-American women, Canter et al. [21] found that 10398A>G polymorphism in the complex I subunit influenced breast cancer susceptibility. In 2010, Czarnecka et al. [31] revealed 23% of the breast cancer cases (10/44) in Polish patients were found to have 10398A>G polymorphism. The authors suggested that 10398A>G polymorphism in the ND3 gene increases the risk of developing breast cancer in patients.
In contrast, recent research revealed that there is no association between the mtDNA 10398A>G and risk of breast cancer. Ismaeel et al. [43] showed that breast cancer risk in Iraqi women had no significantly link with 10398A>G mutation. Their finding did not support the hypothesis that the 10398A>G variant could be considered as a potential risk marker for breast cancer susceptibility in Iraqi women. In more recent case-control study, Jiang et al. [24] reported that the 10398A>G polymorphism was not associated with the risk of breast cancer in Han Chinese women. They also found that there was no correlation between the 10398A>G polymorphism and mtDNA copy number in the cases.
Beside that the 10398A allele variant was also reported to increase the incidence and severity of prostate cancer in African-American men [39]. Moreover, in the cases of thyroid tumors, Yeh et al. [44] analyzed the whole mitochondrial genome in 21 thyroid tumors and detected a somatic 10398A>G mutation in a papillary thyroid carcinoma, a low-grade follicular variant.
The present study showed that mitochondrial ND3 10398A>G mutation was found in 57.8% of brain tumor patients. This is believed to be the first report of studying mitochondrial complex I subunit 10398A>G alteration involving brain tumor of Malaysian patients except there was only one published report on 10398A>G polymorphism in Malaysian population but in breast cancer [41]. Surprisingly, our study demonstrated a quite high 10398A>G mutation rate compared with other previous studies of breast cancer. This high mutation rate may result in accelerated mutation-accumulation due to the presence of many mitochondria in the brain cells. Cells that require high energy demands, such as muscle, brain, CNS, heart, and liver, have many mitochondria. mtDNA is known to be more sensitive to oxidative stress induced damage. Mutations in mtDNA are the source of mitochondrial disease that can affect mostly areas of high energy demand, like the brain cells.
Although most ND3 10398A>G variant has been described in breast cancer patients, to date, it has not been described in brain tumor patients. The exact role of ND3 10398A>G mutation that could lead to brain tumor is unknown due to the limitations of existing data. However, it has been hypothesized that the defect in A10398G together with other genetic and environmental factors, may lead to increased electron leakage and enhanced oxidative stress. This situation may result in generating more ROS which will further contribute to carcinogenesis [10].
By analyzing the correlation ND3 10398A>G mutation status and clinicopathological characteristics, the present study found no significant difference between the ND3 10398A>G mutation group and non-mutation group in histological tumor grade (p<0.05). ND3 10398A>G mutation were detected more frequently in intra-axial tumor compared to extra-axial tumor (p<0.01). However, we did not find a significant relationship between ND3 10398A>G mutation and other clinicopathologic characteristics such as age and gender.
Furthermore, it has been known that IDH mutations are invaluable diagnostic marker/tool that can be used to assist the differential diagnosis in glioma grading [45]. Given the frequent occurrence of IDH1 mutations in gliomas [46], thus we recently reported IDH1 c.395G>A (R132H) mutation (14/40, 35%) in brain tumors tissues, using the same patient cohort [34]. Therefore, in present study, we analysed the association between the ND3 mutation status and IDH1 mutation. We demonstrate that there is no correlation between ND3 mutation and IDH1 mutation.
Based on our results, we did not observe a significant association between ND3 10398A>G mutation status and prognostic factors. The lack of association between the ND3 10398A>G mutation and prognostic factors might have derived from the small number of samples and/or limited follow-up period time, even though we identified a quite high mutational rate of the ND3 10398A>G in brain tumors. Our data suggest that further study with larger sample size (with the extended clinical follow-up and survival rate data) is needed to clarify whether ND3 10398A>G mutation status is correlated to clinical outcome/prognosis in brain tumors. However, it is believed that this ND3 10398A>G mutation could be associated with other mutations and/or factors are capable in initiating and promoting tumorigenesis of the brain.
In conclusion, the present study revealed that mtDNA alterations will enable to be a new biomarker and potential target for identifying individuals at high chance of developing specific cancer types as well as for early screening and detection approach of cancer. We observed a much high frequency of mitochondrial ND3 10398A>G mutation in brain tumors with a significant impact on the high grade of brain tumors especially in intra-axial tumors. We believed that our results are able to contribute as a new finding for the mitochondrial global database. Our data also may provide initial important inputs to how this ND3 10398A>G alteration can be considered to be involved in high-grade or malignant brain tumor progression. Alteration of ND3 10398A>G could be classified as pathogenic mutation in connection with mutations in other mitochondrial or nuclear genes as well as environmental factors in the development of various diseases and cancers. Further research is essentially required to understand the effect and role of the ND3 10398A>G mutation in the brain tumorigenesis. In addition, alteration in the complete of entire coding region of mitochondrial genome in brain tumors in Malaysia patients is also under study.
Acknowledgments
We would like to thank the patients for their participation and cooperation in providing tissue samples and information.
Research partly supported by the Research University Grant (RUI) (1001/PPSP/812110) from Universiti Sains Malaysia and the Fundamental Research Grant Scheme (FRGS) (203/PPSP/6171161) from the Ministry of Higher Education of Malaysia.
References
1. de Robles P, Fiest KM, Frolkis AD, et al. The worldwide incidence and prevalence of primary brain tumors: a systematic review and meta-analysis. Neuro Oncol. 2015; 17:776–783. PMID: 25313193.

2. National Brain Tumor Society. Quick Brain Tumor Facts. Newton, MA: National Brain Tumor Society;2018. Accessed April, 2018. http://braintumor.org/brain-tumor-information/brain-tumor-facts/.
3. Lim GCC, Rampal S, Halimah Y. Cancer incidence in Peninsular Malaysia, 2003–2005. Kuala Lumpur: National Cancer Registry;2008.
4. Goh CH, Lu YY, Lau BL, et al. Brain and spinal tumour. Med J Malaysia. 2014; 69:261–267. PMID: 25934956.
5. Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010; 5:297–348. PMID: 20078222.

6. Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012; 13:780–788. PMID: 23175281.

7. Verschoor ML, Ungard R, Harbottle A, Jakupciak JP, Parr RL, Singh G. Mitochondria and cancer: past, present, and future. Biomed Res Int. 2013; 2013:612369. PMID: 23509753.

8. Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006; 25:4663–4674. PMID: 16892080.

9. Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med. 2002; 4:1–19.

10. Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004; 4:755–762. PMID: 16120430.

11. Singh KK, Kulawiec M. Mitochondrial DNA polymorphism and risk of cancer. Methods Mol Biol. 2009; 471:291–303. PMID: 19109786.

12. Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2011; 1807:620–625. PMID: 21074512.

13. Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila). 2011; 4:638–654. PMID: 21543342.

14. Tseng LM, Yin PH, Yang CW, et al. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer. 2011; 50:800–811. PMID: 21748819.

15. Hsu CC, Lee HC, Wei YH. Mitochondrial DNA alterations and mitochondrial dysfunction in the progression of hepatocellular carcinoma. World J Gastroenterol. 2013; 19:8880–8886. PMID: 24379611.

16. Yang Ai SS, Hsu K, Herbert C, et al. Mitochondrial DNA mutations in exhaled breath condensate of patients with lung cancer. Respir Med. 2013; 107:911–918. PMID: 23507584.

17. Li LH, Kang T, Chen L, et al. Detection of mitochondrial DNA mutations by high-throughput sequencing in the blood of breast cancer patients. Int J Mol Med. 2014; 33:77–82. PMID: 24253185.

18. Lee HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol. 2014; 20:3950–3959. PMID: 24744584.

19. Fang Y, Huang J, Zhang J, et al. Detecting the somatic mutations spectrum of Chinese lung cancer by analyzing the whole mitochondrial DNA genomes. Mitochondrial DNA. 2015; 26:56–60. PMID: 24006865.

20. Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med (Maywood). 2016; 241:1281–1295. PMID: 27022139.

21. Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005; 65:8028–8033. PMID: 16140977.

22. Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007; 249:249–255. PMID: 17081685.

23. Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009; 54:647–654. PMID: 19763141.

24. Jiang H, Zhao H, Xu H, et al. Peripheral blood mitochondrial DNA content, A10398G polymorphism, and risk of breast cancer in a Han Chinese population. Cancer Sci. 2014; 105:639–645. PMID: 24703408.
25. van der Walt JM, Dementieva YA, Martin ER, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004; 365:28–32. PMID: 15234467.

26. Shoffner JM, Brown MD, Torroni A, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993; 17:171–184. PMID: 8104867.

27. Kösel S, Grasbon-Frodl EM, Mautsch U, et al. Novel mutations of mitochondrial complex I in pathologically proven Parkinson disease. Neurogenetics. 1998; 1:197–204. PMID: 10737123.

28. van der Walt JM, Nicodemus KK, Martin ER, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003; 72:804–811. PMID: 12618962.

29. Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007; 67:4687–4694. PMID: 17510395.

30. Setiawan VW, Chu LH, John EM, et al. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet Cytogenet. 2008; 181:16–19. PMID: 18262047.

31. Czarnecka AM, Krawczyk T, Zdrozny M, et al. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res Treat. 2010; 121:511–518. PMID: 19266278.

32. Mohamed Yusoff AA, Mohd Nasir KN, Haris K, et al. Detection of somatic mutations in the mitochondrial DNA control region D-loop in brain tumors: the first report in Malaysian patients. Oncol Lett. 2017; 14:5179–5188. PMID: 29098023.

33. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109. PMID: 17618441.

34. Mohamed Yusoff AA, Zulfakhar FN, Sul'ain MD, Idris Z, Abdullah JM. Association of the IDH1 C.395G>A (R132H) mutation with histological type in Malay brain tumors. Asian Pac J Cancer Prev. 2016; 17:5195–5201. PMID: 28125199.
35. Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992; 61:1175–1212. PMID: 1497308.

36. Dimauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005; 37:222–232. PMID: 16019721.

38. Grzybowska-Szatkowska L, Slaska B. Mitochondrial DNA and carcinogenesis (review). Mol Med Rep. 2012; 6:923–930. PMID: 22895648.
39. Mims MP, Hayes TG, Zheng S, et al. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2006; 66:1880. author reply 1880-1. PMID: 16452251.

40. Sultana GNN, Rahman A, Karim MM, Shahinuzzaman ADA, Begum R, Begum RA. Breast cancer risk associated mitochondrial NADHdehydrogenase subunit-3 (ND3) polymorphisms (G10398A and T10400C) in Bangladeshi women. J Med Genet Genomics. 2011; 3:131–135.
41. Tengku Baharudin N, Jaafar H, Zainuddin Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. Malays J Med Sci. 2012; 19:36–42.
42. Yu Y, Lv F, Lin H, et al. Mitochondrial ND3 G10398A mutation: a biomarker for breast cancer. Genet Mol Res. 2015; 14:17426–17431. PMID: 26782384.

43. Ismaeel HM, Younan HQ, Zahid RA. Mitochondrial DNA A10398G mutation is not associated with breast cancer risk in a sample of Iraqi women. Curr Res J Biol Sci. 2013; 5:126–129.
44. Yeh JJ, Lunetta KL, van Orsouw NJ, et al. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000; 19:2060–2066. PMID: 10803467.

45. Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014; 15:e395–e403. PMID: 25079102.

46. Wang XW, Ciccarino P, Rossetto M, et al. IDH mutations: genotype-phenotype correlation and prognostic impact. Biomed Res Int. 2014; 2014:540236. PMID: 24877111.
Fig. 1
Agarose gel electrophoresis of mitochondrial ND3 PCR products. Amplification products of the expected size of 422-bp was detected in 2% agarose gels. Lane M, 100 bp DNA marker; Lane 1, negative control; Lane 2–7, tumor samples.
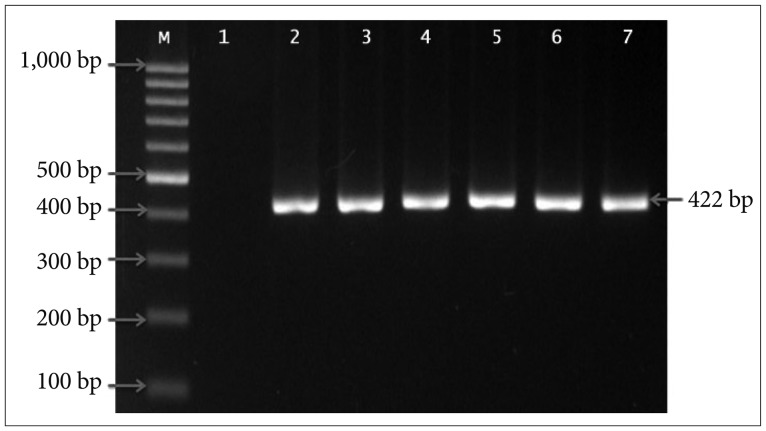
Fig. 2
PCR-RFLP analysis of ND3 (A10398G) mutations in brain tumor patients. Fragment of ND3 gene digested with DdeI (Hpy-F3I) enzyme. Lane M, 100 bp DNA marker; Lane 1, undigest product (422-bp); Lane 2, normal sample; Lane 3–7, tumor samples. Three bands at 213-bp, 130-bp & 80-bp indicate a wild type. Four bands at 175-bp, 130-bp, 80-bp & 38-bp indicate a homozygous mutation. Five bands at 213-bp, 175-bp, 130-bp, 80-bp & 38-bp indicate a heterozygous mutation.
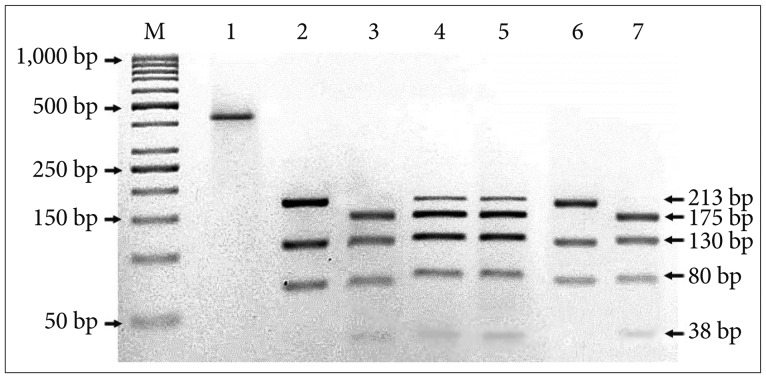
Fig. 3
Representative sequencing results showing example of mitochondrial ND3 mutations in brain tumor tissue sample. Chromatograms show a mutation of A to G transition at nucleotide position 10398.
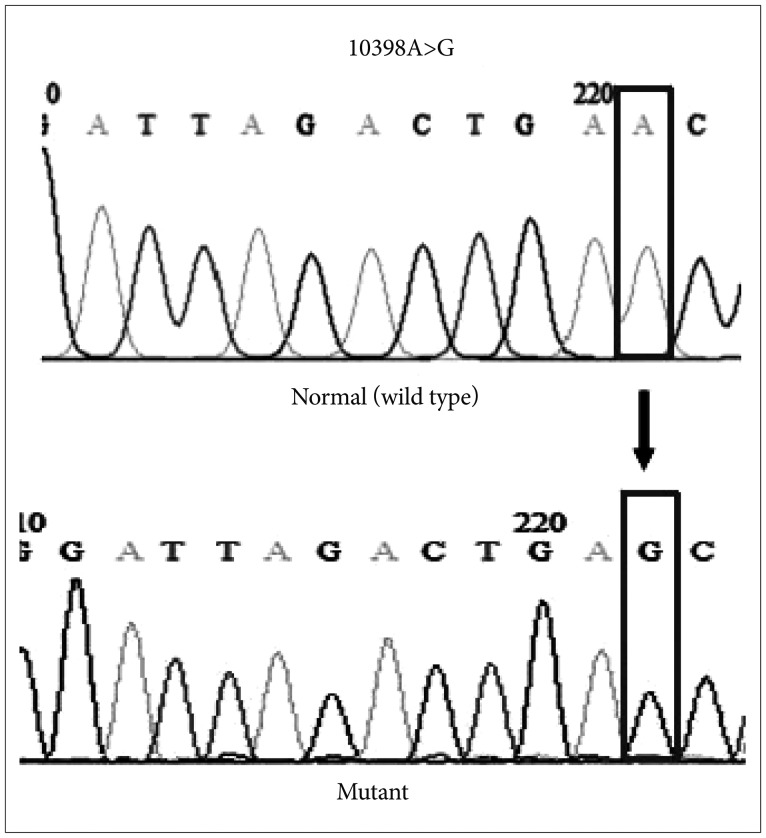
Fig. 4
The pie charts depict the percentages of ND3 10398A>G mutations in brain tumor patients. A: Mutation vs. wild type. B: Heterozygous mutations vs. homozygous mutations.
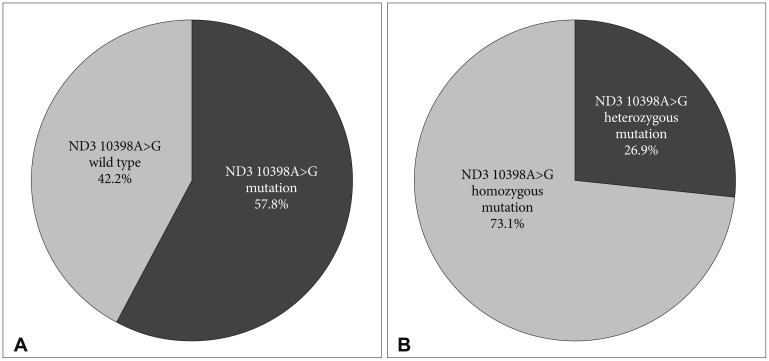
Table 1
Clinicopathologic profiles of the brain tumor cases in the study
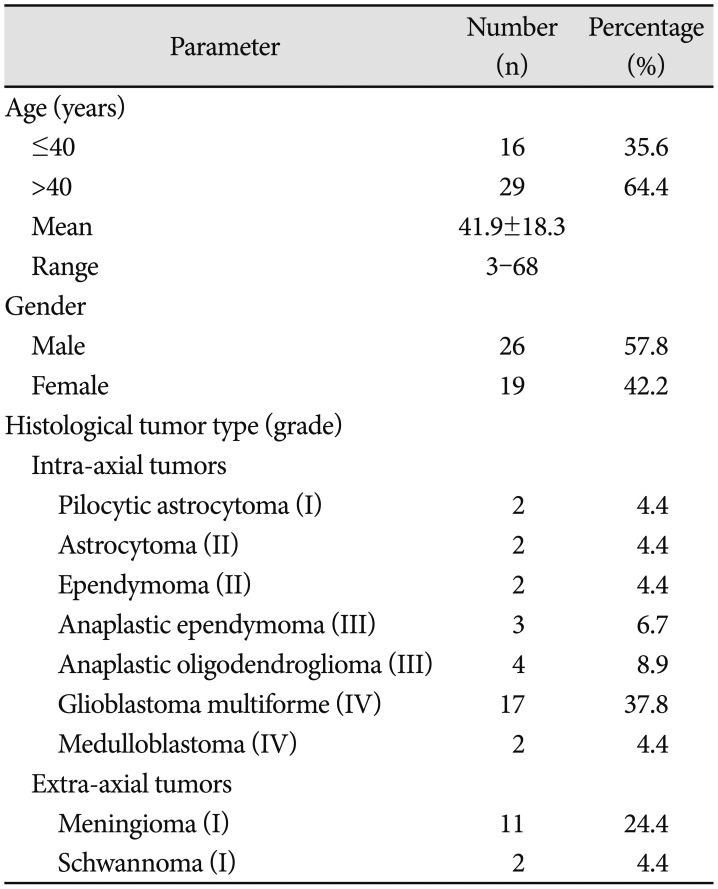
Table 2
Correlation of ND3 10398A>G mutations status with clinicopathological parameters in brain tumor
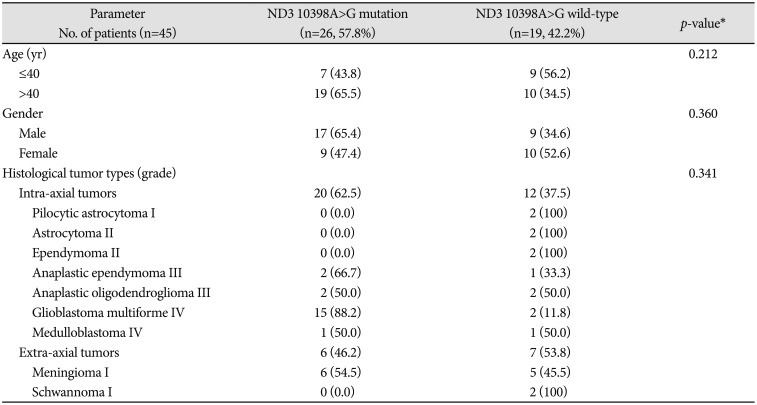
Table 3
IDH mutation association of ND3 10398A>G mutation status

| Total (n=40) | ND3 10398A>G mutation | p-value* | ||
|---|---|---|---|---|
| ND3mut | ND3wild | |||
| IDH1 c.395G>A (R132H) mutation† | ||||
| IDH1mut | 14 | 8 | 6 | >0.999 |
| IDH1wild | 26 | 14 | 12 | |
*Fisher's exact test was used, †Data was taken from Mohamed Yusoff et al. Asian Pac J Cancer Prev 2016;17:5195–201 [34].




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download