Abstract
Background
To investigate the molecular basis for invasion of malignant gliomas, proteomic analysis approach was carried out using two human glioma cell lines, U87MG and U343MG-A that demonstrate different motility and invasiveness in in vitro experiments.
Methods
High-resolution two-dimensional gel electrophoresis and matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry analysis were performed.
Gliomas are a heterogenous group of neoplasms derived from various glial cells and account for 40-50% of all intracranial tumors. Glioblastoma multiforme (GBM), the most malignant type of glioma (WHO grade IV), is highly invasive and tend to diffusely infiltrate into the surrounding normal brain parenchyma. These characteristics prevent the tumor cells to reside within the margins of the therapeutic resection. Although the mechanisms of invasion in gliomas have been investigated in different human samples and experimental studies, there is a need for a novel panel of biomarkers that characterize each invasive phenotype.
Traditionally, gene expression profiles have identified many genes with distinct expression patterns among different histological types and grades of gliomas [1,2,3]. However, biological systems comprise protein components resulting from transcriptional and post-transcriptional control, post-transcriptional modifications, and shifts in proteins among the different cellular compartments. These properties cannot be analyzed by microarray systems at the RNA level, whereas proteomic analysis such as high-resolution two-dimensional gel electrophoresis (2DGE) and mass spectrometry (MS) separate and identify the targeted proteins [4,5]. Proteomics has recently emerged as a new field of protein science that allows for high-throughput profiling of expressed proteins at cellular and subcellular levels. This proteomic-based analysis promises new insights into the cellular mechanisms involved in tumor invasion and is likely to result in the discovery of novel diagnostic and therapeutic targets.
In our previous study, different migration ability between human GBM cell lines was confirmed by simple scratch migration assay [6]. Furthermore, we identified some genes possibly related to the high motility in GBM cell lines using differentially display-polymerase chain reaction (PCR) by reverse transcription-PCR and Northern blot analysis [6], and have continued to reveal the downstream mechanism for these genes [7,8,9,10].
In this study, based on the migration/invasion ability of the high motile cell line (U87MG) and less motile cell line (U343-MG-A), we examined these cell lines using a proteomics approach, including the use of 2DGE and matrix assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) analysis, to compare the proteomic profiles of these two cell lines with different motility.
U87MG and U343MG-A (the American Type Culture Collection, Rockville, MS, USA) was maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, Carlsbad, CA, USA) media supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) under 5% CO2 at 37℃.
2×104 cells of U87MG and U343MG-A in 0.35 mL of serum-free DMEM were seeded into the upper chamber of a 10-well chemotaxic chamber (Neuro Probe, Gaitherburg, MD, USA) and complete DMEM was placed in the lower chamber, and a Matrigel-coated membrane was inserted between the two chambers. After 24 hours incubation at 37℃, membranes were fixed and stained using a hemacolor rapid staining kit (Merck, Darmstadt, Germany), the cells from 5 random microscopic fields (220× magnification) were counted. Same experiment was repeated 3 times, independently.
For complete inhibition of cell proliferation, 5 mM hydroxyurea was added to the culture media for 24 hours. The cultures were then scratched with a single-edged razor blade, washed twice with PBS, and placed in media containing hydroxyurea. After 46 hours incubation, the cultures were immersed in methanol and stained with 0.1% toluidine blue. Three microscopic fields were evaluated for each wound injury. The number of cells that traversed the scratched area and the maximum distance traveled were determined in each field and averaged for each injury. These experiment were repeated three times.
For proteome analysis, both cell lines were passed when an approximate 80% confluence was reached, then each cell line was homogenized in lysis buffer consisting of 7 M urea, 2 M thiourea, 2% CHAPS, 1% dithiothreitol (DTT), 0.5 mM EDTA, and 1× protease inhibitors. The samples were centrifuged at 14000 g for 20 min at 18-20℃. Supernatants were then separated and used for 2DE after protein quantification using the BSA assay (Bio-Rad Laboratories, Munich, Germany).
The first dimension of 2DE was performed on an immobilized pH gradient (IPG) strip (Amersham Biosciences, Amersham Pl, UK). Linear pH 3-10 IPG strips (18 cm) were rehydrated overnight at room temperature in rehydrating buffer (8 M urea, 1% DTT, 2% CHAPS, and 0.5% IPG buffer). Sample of 300 mg was applied during rehydration. The first dimension was run for 53500 Vh at 20℃ using the following conditions: 500 V for 1 h, 1000 V for 1 h; and 8000 V for 6 h and 30 min. Next, gels were equilibrated for 30 min in a equilibration buffer I containing 50 mM Tris-Cl, 6 M urea, 30% glycerol, 2% sodium dodesyl sulfate (SDS) and 0.1% DTT and subsequently equilibration buffer II containing 50 mM Tris-Cl, 6 M urea, 30% glycerol, 2% SDS and 0.25% iodoacetic acid. The second dimension was run on Ettan DALT II system (Amersham Biosciences, Amersham Pl, UK). An 8-15% gradient acrylamide gel was used for the second dimensional gel electrophoresis. The IPG strips were placed on the surface of the second dimensional gel, and then were sealed with 0.5% agarose in SDS electrophoresis buffer containing 25 mM Tris base, 192 mM glycine and 0.1% SDS. The gels were placed into Ettan DALT II system chamber containing 1% SDS electrophoresis buffer. The gels were run overnight at 100 V until the dye front reached the bottom of the gel.
Proteins were visualized using coomassie blue staining, in which the intensity of the protein spot is correlated with the quantity of protein present. The gels were fixed at room temperature in 12% tri-chloroacetic acid for 60 min and washed twice in distilled water for 15 min. Next, the gels were staining overnight with 160 mL of staining solution (0.1% coomassie G-250 in 2% H3PO4, 10% ammonium sulfate) plus 40 mL of methanol, and washed for 1 to 3 min in 0.1 mol/L Tris, H3PO4 buffer, ph 6.5, then rinse for 1 min in 25% aqueous methanol. The gels were then stabilized the protein-dye complex in 20% aqueous ammonium sulfate.
Coomassie blue-stained gels were analyzed to select spots that appeared consistently between two samples. Selection of the spots was based on volume and density as determined using Phoretix 2D Advanced software (Version 5.01; Nonlinear Dynamic, Newcastle, UK). The selected proteins were excised under a laminar flow hood using a scalpel and then processed for protein digestion. An accelerated digestion protocol was adopted as described by Havlis et al. [11]. Briefly, the Coomassie blue-stained protein spots were excised, destained, and dehydrated in ammonium bicarbonate and acetonitrile (ACN) for 20 min. The solution was then aspirated, and gel pieces were brought to complete dryness in a SpeedVac. Gel pieces were rehydrated with a freshly prepared solution of sequence-grade modified trypsin (20 ng/µL in 25 mM ammonium bicarbonate buffer) at 4℃ for 60 min and then incubated at 55℃ for 60 min. The tryptic peptides were extracted with 10 µL of 1% formic acid aqueous solution by sonication in a water bath for 15 min. The resulting samples were stored at -80℃ until analyzed by MS.
Tryptic-digest peptides were concentrated and desalted using GELoader microcolumns packed with POROS 20 R2 resin (Applied Biosystems, Foster City, CA, USA). The peptides were then eluted with 1 µL matrix solution (70% ACN, 0.1% trifluoroacetic acid). The eluted peptides were then spotted on to a stainless steel target plate and peptide masses were determined using MALDI-TOFMS and an Applied Biosystems 4700 proteomics analyzer (Applied Biosystems). The unit was calibrated using external mass standards (Calmix 1 and 2; Applied Biosystems). Peptide masses were compared with the theoretical peptide masses of all human proteins in the NCBI database using Mascot software (Matrix Sciences, London, UK). A Mascot probability-based score for protein identification of greater than 50 was considered acceptable. The identified proteins were also examined for matched peptides and sequence coverage: five or more matched peptides or 30% or greater sequence coverage was considered acceptable.
Quantitative data were collected for invasion assay, scratch test, and migration assay, as described above. These experiments were performed in triplicate and repeated three times. The observed effects was evaluated by the Mann-Whitney U test using SPSS version 20.0 software program for Windows (SPSS Inc., Chicago, IL, USA). The level of significance was set at p<0.05.
In invasion assay, the number of invasive cells (mean±standard deviation) was 179.3±15.1 in U87MG and 25.5±7.5 in U343MG-A. There was a significant difference in both cell lines (p<0.001), as shown in Fig. 1. The number of migrated cells was 473±112.4 in U87MG and 80.2±13.5 in U343MG-A. This difference was also statistically significant (p<0.001), as shown in Fig. 2.
Protein expression maps of U87MG and U343MG-A were generated using 2DGE. The 2DGE gel images of these cell lines demonstrated similar patterns of protein expression, due to the genetic similarity between them (Fig. 3). With Phoretix 2D Advanced software for 2DGE gel analysis, numerous well-stained protein spots were detected. Only nine spots revealed significant difference in spot density and volume. Compared to the spot intensity and volume of U343MG-A, spots 509, 561, 566, 613, 618 were up-regulated and spots 171, 274, 342, 475 were down-regulated in U87MG cells. The result of different expressed protein spots was summarized with circles and spot numbers as shown in Fig. 4.
We analyzed differentially expressed protein spots, followed by excision from the gel, using MALDI-TOFMS and database searching. The 9 identified proteins in spots are summarized in Table 1. The higher score or greater number of matched peptide/sequence coverage indicated the better probability. Proteins that were up-regulated in U87MG included one proteinase protein (cathepsin D), two structural proteins (eukaryotic translation inhibition factor 6 and nicotinamide phosphoribosyl transferase), one RNA-binding protein (ribonuclease inhibitor 1) and one metabolic enzyme protein (transglutaminase 2). Down-regulated proteins in U87MG were one metabolic enzyme protein (glyceraldehyde-3-phosphate dehydrogenase), one ATP-binding protein (autophagy related 7), one F-box protein (F-box only protein 47) and one signal transduction protein (calcium/calmodulin-dependent phosphodiesterase 1A).
The current study describes the proteome maps of human GBM cell lines, U87MG and U343MG-A. Considering the fact that U87MG is a more motile cell line than U343MG-A, some of the different expressed proteins may be related with motility or invasion in glioma cell lines. Two cell lines showed similarities in the protein expression patterns on the 2DGE gel images, reflecting that there is closeness in the genetic background. This point was observed in another proteomic analysis using sibling glioma cell lines, J3T-1 and J3T-2, with different invasion property [12]. Among the only 18 protein spots that are differently expressed and identified, annexin A2 was focused on as a candidate invasion-related protein, involved in angiogenesis-dependent patterns in gliomas.
In our study, one of the proteins identified, cathepsin D, was expressed at higher levels in U87MG than in U343MG-A. Cathepsin D is normally localized within the lysosomes and involved in degradation of proteins and processing of precursor proteins [13]. It plays an essential role in tumor progression, angiogenesis and apoptosis, in addition to the protein turnover process [14,15,16]. In a recent investigation, cathepsin D prevented oxidative stress-induced cell death through the activation of autophagy in cancer cells. This result suggests that inhibition of autophagy may be a novel therapeutic strategy for human cancers which express cathepsin D [17]. Fukuda et al. [18] suggested that cathepsin D may be a potential serum marker for the prediction of aggressive nature of human gliomas. In neuroblastomas, cathepsin D has an important role in anti-apoptotic signaling and is a novel mechanism for the chemo-resistance [15].
Based on proteomic analysis, Iwadate et al. [19] found that 37 proteins differentially expressed based on the histologic grading of human gliomas. Among these proteins, cathepsin D was increased in high-grade gliomas and may be one of novel biomarkers for survival prediction and rational target for anti-glioma therapy. Recent investigations revealed that cathepsin D was confirmed to be up-regulated in high-grade classification and possibly related to acquired temozolomide resistance in human malignant gliomas [20]. Liu et al. [21] proposed that the inhibition of lysosome cathepsin D exocytosis from glioma cells plays a pivotal modulatory role in their migration and invasion. Furthermore, promotion of proliferation and invasion, and suppression of apoptosis in glioma cells induced by Rab27a were exerted by increasing cathepsin D and regulated by miR-24 [22]. Our study also implicates the possible role of cathepsin D in glioma motility and invasion. However, further studies using knock-down or stable transfection are necessary to reveal the exact role and mechanism of cathepsin D in glioma invasion.
Transglutaminase 2, one of the up-regulated proteins in U87MG in our experiment, has not been widely investigated in glioma research. However, recent papers proposed that this protein is possibly related to drug resistance in glioma cells [23]. Although there have been some papers showing the inhibitory role of ribonuclease inhibitor 1 on cancer growth and metastasis [24], there have been no past studies addressing the role of ribonuclease inhibitor 1 in gliomas. Nicotinamide phosphoribosyltransferase has been known as one of the key enzymes in salvaging pathway for the synthesis of nicotinamide adenine dinucleotide, involved in cell metabolism and proliferation [25]. Considering the high metabolic demand in malignant gliomas, the inhibition of nicotinamide phosphoribosyltransferase may reduce nicotinamide adenine dinucleotide to inhibit the growth of glioblastomas [26]. However, there is no previous study on the relation between nicotinamide phosphoribosyltransferase and glioma invasion. This has not been investigated in the role of eukaryotic translation inhibition factor 6 in gliomas.
In conclusion, using proteomic analysis for two glioma cell lines with different motility and invasiveness, some proteins were differently expressed. Although there was no protein directly connected to tumor cell motility among them, cathepsin D seemed be one of the possible proteins which are related to glioma invasion.
Acknowledgement
This study was supported by a grant (CRI 09021-1) of Chonnam National University Hospital Research Institute of Clinical Medicine.
References
1. Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002; 415:436–442. PMID: 11807556.

2. Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001; 61:6885–6891. PMID: 11559565.
3. Sallinen SL, Sallinen PK, Haapasalo HK, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000; 60:6617–6622. PMID: 11118044.
4. Celis JE, Gromov P, Ostergaard M, et al. Human 2-D PAGE databases for proteome analysis in health and disease: http://biobase.dk/cgi-bin/celis. FEBS Lett. 1996; 398:129–134. PMID: 8977092.
5. Godovac-Zimmermann J, Brown LR. Perspectives for mass spectrom-etry and functional proteomics. Mass Spectrom Rev. 2001; 20:1–57. PMID: 11223909.

6. Ryu HH, Jung S, Sun HS, et al. Screening for motility-associated genes in malignant astrocytoma cell lines. J Neurooncol. 2007; 82:125–131. PMID: 17048098.

7. Park SG, Jung S, Ryu HH, et al. Role of 14-3-3-beta in the migration and invasion in human malignant glioma cell line U87MG. Neurol Res. 2012; 34:893–900. PMID: 22925547.

8. Ryu HH, Jung S, Jung TY, et al. Role of metallothionein 1E in the migration and invasion of human glioma cell lines. Int J Oncol. 2012; 41:1305–1313. PMID: 22843066.

9. Jung TY, Jung S, Ryu HH, et al. Role of galectin-1 in migration and invasion of human glioblastoma multiforme cell lines. J Neurosurg. 2008; 109:273–284. PMID: 18671640.

10. Jung S, Paek YW, Moon KS, et al. Expression of Nm23 in gliomas and its effect on migration and invasion in vitro. Anticancer Res. 2006; 26:249–258. PMID: 16475705.
11. Havlis J, Thomas H, Sebela M, Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem. 2003; 75:1300–1306. PMID: 12659189.
12. Maruo T, Ichikawa T, Kanzaki H, et al. Proteomics-based analysis of invasion-related proteins in malignant gliomas. Neuropathology. 2013; 33:264–275. PMID: 23116197.

13. Diment S, Martin KJ, Stahl PD. Cleavage of parathyroid hormone in macrophage endosomes illustrates a novel pathway for intracellular processing of proteins. J Biol Chem. 1989; 264:13403–13406. PMID: 2760027.

14. Yin L, Stearns R, González-Flecha B. Lysosomal and mitochondrial pathways in H2O2-induced apoptosis of alveolar type II cells. J Cell Biochem. 2005; 94:433–445. PMID: 15534871.

15. Sagulenko V, Muth D, Sagulenko E, Paffhausen T, Schwab M, Westermann F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis. 2008; 29:1869–1877. PMID: 18566016.

16. Berchem G, Glondu M, Gleizes M, et al. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002; 21:5951–5955. PMID: 12185597.

17. Hah YS, Noh HS, Ha JH, et al. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012; 323:208–214. PMID: 22542809.

18. Fukuda ME, Iwadate Y, Machida T, et al. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005; 65:5190–5194. PMID: 15958563.

19. Iwadate Y, Sakaida T, Hiwasa T, et al. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004; 64:2496–2501. PMID: 15059904.

20. Sun S, Wong TS, Zhang XQ, et al. Protein alterations associated with temozolomide resistance in subclones of human glioblastoma cell lines. J Neurooncol. 2012; 107:89–100. PMID: 21979894.

21. Liu Y, Zhou Y, Zhu K. Inhibition of glioma cell lysosome exocytosis inhibits glioma invasion. PLoS One. 2012; 7:e45910. PMID: 23029308.

22. Wu X, Hu A, Zhang M, Chen Z. Effects of Rab27a on proliferation, invasion, and anti-apoptosis in human glioma cell. Tumour Biol. 2013; 34:2195–2203. PMID: 23553027.

23. Dyer LM, Schooler KP, Ai L, et al. The transglutaminase 2 gene is aberrantly hypermethylated in glioma. J Neurooncol. 2011; 101:429–440. PMID: 20596752.

24. Yao X, Li D, Xiong DM, Li L, Jiang R, Chen JX. A novel role of ribonuclease inhibitor in regulation of epithelial-to-mesenchymal transition and ILK signaling pathway in bladder cancer cells. Cell Tissue Res. 2013; 353:409–423. PMID: 23703635.

25. Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005; 280:36334–36341. PMID: 16118205.

26. Zhang LY, Liu LY, Qie LL, et al. Anti-proliferation effect of APO866 on C6 glioblastoma cells by inhibiting nicotinamide phosphoribosyltransferase. Eur J Pharmacol. 2012; 674:163–170. PMID: 22119381.

Fig. 1
Matrigel invasion assay. The number of invasive cells (mean±SD) was 179.3±15.1 in U87MG (A) and 25.5±7.5 in U343MG-A (B). U87MG cell line was more invasive than U343MG-A with a statistical significance (p<0.001) (C). SD: standard deviation.
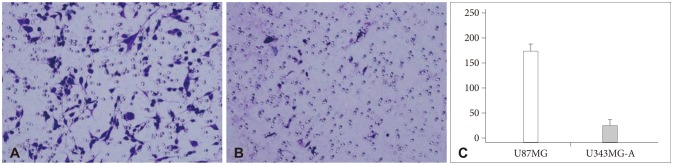
Fig. 2
Scratch test. The number of migrated cells (mean±SD) was 473±112.4 in U87MG (A) and 80.2±13.5 in U343MG-A (B). U87MG cell line was more motile than U343MG-A with a statistical significance (p<0.001) (C). SD: standard deviation.

Fig. 4
Protein spots differentially expressed between U343MG-A and U87MG. Note that the spot intensity & volume of U87MG are described compared to those of U343MG-A.
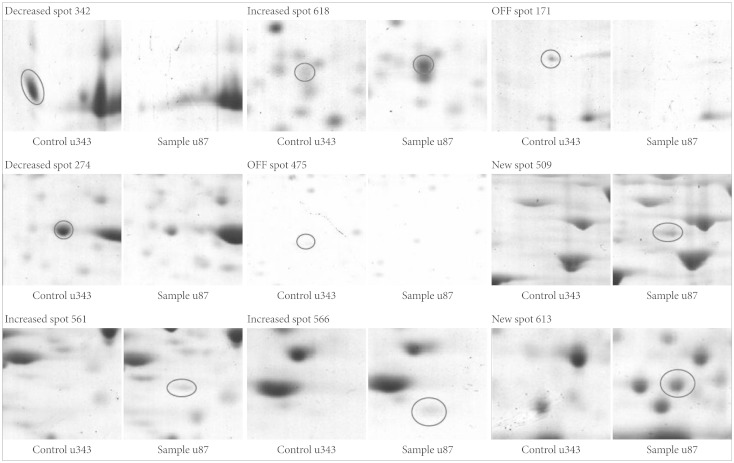
Table 1
List of differentially expressed protein spots in U87MG and U343MG-A
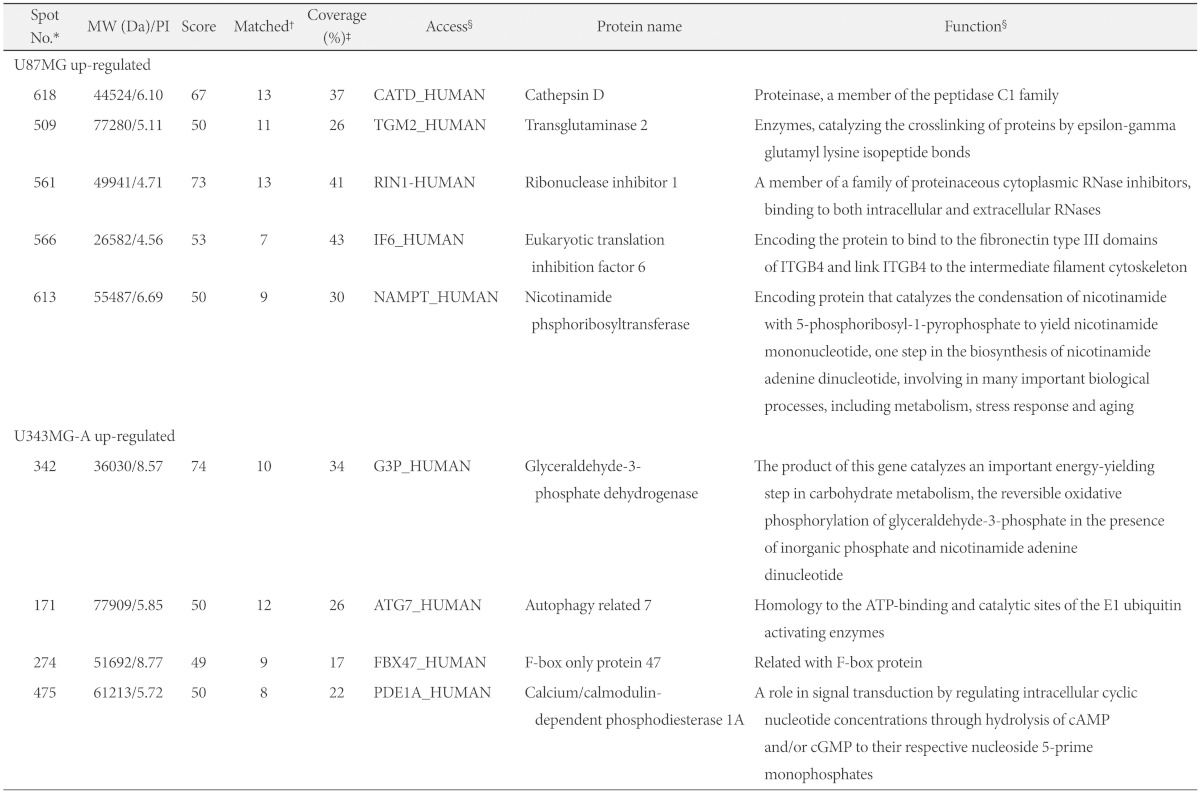
*Protein spot number corresponded to the 2DGE image in Fig. 3A, †Number of matched amino acid, ‡Percent of matched sequence, §From PubMed protein database (http://www.ncbi.nlm.nih.gov/protein). Da: daltons, No.: number, MW: molecular weight, PI: theoretical isoelectric point




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download