This article has been
cited by other articles in ScienceCentral.
Abstract
To develop the large scale serological assay for severe fever with thrombocytopenia syndrome virus (SFTSV) infection, we evaluated two different enzyme-linked immunosorbent assay (ELISA) methods using nucleocapsid protein (NP) and Gn proteins of CB1 (genotype B) SFTSV strains. The NP-based ELISA tests showed more sensitive with broad cross-reactivity between two different genotype A and B strains compared with those of Gn-based ELISA tests. However, Gn-based ELISA showed more genotype specificity and specificity. These result suggested that NP-based ELISA test could be applicable for general sero-prevalence studies of SFTSV infections, while Gn-based ELISA could be applicable for a certain specific genotype sero-prevalence study.
Keywords: Severe fever with thrombocytopenia syndrome virus, Enzyme-linked immunosorbent assay, NP protein, Gn protein
Severe fever with thrombocytopenia syndrome (SFTS) is a newly emerged tick-mediated infectious disease in humans which belongs to the family
Bunyaviridae and has three single-stranded RNA segments consisting of S, M, and L segments [
12]. Since SFTS infectious disease was first reported in China, 2009 [
3], severe fever with thrombocytopenia syndrome virus (SFTSV) has been also reported in South Korea (2013) and Japan (2013) [
45]. Although the name of SFTS is derived from the clinical symptoms of the disease, severe fever and thrombocytopenia, the clinical manifestation of SFTS is nonspecific and difficult to distinguish from those of human granulocytic anaplasmosis, leptospirosis,
Orientia tsutsugamushi infection, and hemorrhagic fever with renal syndrome [
6]. The SFTSV was also isolated from the domesticated animal farms with high seroprevalence where SFTS human patients were previously reported with high genetic homology [
78]. Further, recent studies showed high heritage diversity of SFTSV strains in East Asia (genotypes A to F) [
9]. Therefore, the sensitive and specific serological diagnosis methods are needed to understand the sero-prevalence of multiple genotypes of SFTSV strains.
To detect anti-SFTSV antibodies in sera from infected hosts, immunofluorescence assay (IFA) is routinely used as a reference test for high sensitivity, but it has nonspecific interactions with various viral antigens that cause poor specificity [
10]. Further, this IFA method has several disadvantages, such as the need to handle the live virus, which requires special facilities with high-level biosafety equipment and laborious to apply for large numbers of serum samples [
11]. Due to these limitations, in-house indirect enzyme-linked immunosorbent assays (ELISA) and double-antigen sandwich ELISA have been developed to detect immunoglobulins with the nucleocapsid protein (NP) of SFTSV [
121314]. However, limited ELISA methods are utilized the Gn protein which is the main surface glycoprotein to produce the neutralizing antibody against SFTSV infection [
15].
In this study, we developed the antibody capture ELISA method with Escherichia coli expressed NP and Gn proteins of CB1 (genotype B) Korean strain and compared their sensitivity and specificity with SFTSV confirmed human and experimentally infected ferret sera including each of negative sera. To evaluate the sensitivity and specificity of our ELISA, we adapted the IFA test as a reference analysis. To this end, two previously reported CB1 (genotype B) and CB2 (genotype A) SFTSV strains [
16] were used for antigen preparedness and each genotype specific antibody generations. Briefly, each genotype specific SFTSV positive sera were generated from the immunization of inactivated CB1 (genotype B) and CB2 (genotype A) SFTSV strains in SFTSV negative ferrets (n=3). To eliminate the any possible growth of SFTSV in immunized ferrets, SFTSV specific real time reverse transcriptase-polymerase chain reaction using the white cells of the ferrets every 3 dpi. No virus was detected from all the time points (data not shown). After the second vaccination, whole serums were collected and combined from each ferret and used as a CB1- or CB2-positive reference antibody sera. All animal experiments were approved by the Medical Research Institute, a member of Laboratory Animal Research Center of Chungbuk National University (LARC) (approval number: CBNUA-986-16-01) and were conducted in strict accordance and adherence to relevant policies regarding animal handling as mandated under the Guidelines for Animal Use and Care of the Korea Center for Disease Control and Prevention (KCDC). The handling of viruses was performed in an enhanced biosafety level 3 (BSL3) containment laboratory as approved by the KCDC (KCDC-14-3-07).
To use the ELISA coating antigens,
E. coli expressed recombinant NP and Gn proteins of CB1 strain were purified as described in elsewhere [
1718]. Then, 100 ng per well of purified each protein was coated onto Polysorp ELISA plates (Nunc, Rochester, NY, USA) for 16 hours at 4℃. After the blocking to prevent non-specific binding, 10-fold diluted serum were incubated on coated plates for 5 hours and horseradish peroxidase peroxidase (HRP)–conjugated anti body (1:1,000) were incubated for 2 hours. Following the washing with phosphate buffered saline with Tween-20 (PBST), ortho-phenylenediamine peroxidase substrate (Sigma, St. Louis, MO, USA) was added as the HRP substrate. The color development was terminated with 1N H
2SO
4. The optical density (OD) was measured spectrophotometrically at 490 nm. For comparison study, the IFA was performed as a slightly modified previously described [
19]. Briefly, Vero E6 cells in 6-well plates were infected with 1 mL of 1×10
3 TCID
50/mL of CB1 and CB2 SFTSV strains for 2 hours at 37℃ and incubated with 3 days. The infected cells were fixed with 80% acetone solution. The serially diluted field serum samples were incubated with fixed cells for 3 hours at 37℃, and the fluorescence was detected using fluorescein isothiocyanate–labeled secondary antibodies. The fluorescence was observed using an Olympus IX 71 (Olympus, Tokyo, Japan) microscope and DP controller software to capture images.
To assess significant differences in ELISA between positive and neagative samples were compared. Asterisks indicate the statistical significance between negative and other sampels determined by t test. All statistical analyses were performed with using GraphPad Prism version 5.00 for Windows (GraphPad Software, La Jolla, CA, USA).
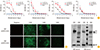
To evaluate our NP- and Gn protein–based ELISAs, we used our laboratory generated CB1 and CB2 genotype specific SFTSV-positive ferret sera which were confirmed antibody titers by IFA test. NP-based ELISA test against CB1 strain anti-sera showed 1.01–1.18 OD value and increased up to 1.64–1.92 OD value by boosting immunization (
Fig. 1A). Further, Gn-based ELISA test also showed relative high OD value 1.04 to 1.49 and increased to 1.07–1.98 by boosting immunization against homologous CB1 strain. In heterologous genotype assay with CB2 (genotype A) anti-sera, NP-based ELISA result showed almost similar OD values as 1.00-1.14 OD value and increased up to 1.54–1.82 OD value by boosting immunization. However, Gn-base ELISA showed relatively low OD values as 0.80–1.08 compared with those of homologous genotype assay even by boosting immunization. However, mock (PBS) immunized ferret sera showed low OD values as 0.15 to 0.27 in both assays. The IFA results showed that CB2 ferret anti-sera could detect the both CB1 (genotype B) and CB2 (genotype A) viruses in Vero E6 cell although the higher intensity of IFA was observed in homologous virus and the detection limit was 1:400. (
Fig. 1B). To confirm the antigenic difference between SFTSV genotype A and B strains, we adapted Western blot assay with purified CB1 and CB2 strains by ultracentrifugation (25,000 rpm, 3 hours, XE-90, Beckman Coulter, Brea, CA, USA). Western blot assay showed that purified CB1 and CB2 viruses showed different pattern of positive bands against Gn/Gc proteins although the NP specific bands were detected in both antibodies (
Fig. 1C). The CB1 immunized sera could detect the specific Gn proteins of CB1 virus, but it only detects unclear Gn bands of CB2 strains (
Fig. 1C, left panel). Further, reversed pattern was detected in CB2 anti-sera test (
Fig. 1C, right panel). These result suggested that there are antigenic differences between genotype A and B SFTSV strains.
In addition, to evaluate the Gn-and NP-based ELISAs with human sera, seven confirmed SFTSV and 20 negative human sera by IFA were evaluated for both ELISA assay. These SFTSV suspected sera were collected from Chungbuk National University hospital to identify infection of SFTSV with patients' permissions. The diagnosis and related research of SFTSV suspected patients' specimens were authorized by Ethics Committee of the Faculty of Medicine at Chungbuk National University (approval No. IRB 2017-05-002-001). NP-based ELSIA results showed 1.08 to 1.29 OD in seven SFTSV positive sera (
Fig. 2A). However, Gn-based ELISA showed relatively broad ranges of OD values from 0.74 to 1.40 against SFTSV positive sera although all negative sera showed low basal levels of OD values as 0.20 to 0.42 (
Fig. 2B). Interestingly, the P1 and P2 sera showed higher OD values in Gn-based ELISA compared with NP-based ELISA tests, while three positive samples (P5, P6, and P7) showed significantly lower OD values in Gn-based ELISA than those of NP-based ELISA (1.15–1.18 vs. 0.91–0.95) (
Fig. 2A). These results suggested that multiple genotypes of SFTSV strains are co-circulating in South Korea and they might be serologically differentiated by surface glycoproteins such as Gn protein. Further, NP-based ELISA showed broader cross reactivity between different genotypes of SFTSVs, while Gn-based ELISA could support to each Genotype specific antibody responses.
In this study, we established NP- and Gn-based ELISA methods with
E. coli expressed genotype B (CB1) which is most prevalence strain in Korea [
16]. Both NP- and Gn-based ELISA methods showed relatively well agreement against homology SFTSV CB1 strain. Further Gn-based ELISA showed more rapid and high OD values against the homologous CB1 immunized ferret sera, but delayed (needed boosting) and relatively low OD value against the heterologous CB2 immunized ferret sera. Comparative results showed that the NP-based ELISA method was more advantageous for evaluating general sero-prevalence because of its higher cross-reactivity between SFTSV genotypes. However, Gn-based ELISA method could be applicable to establish genotype specific diagnosis methods. Taken together, we believed that the NP- and Gn-based ELISA methods established in this study can be useful for the measurement of infection rates in animals as well as humans.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download