This article has been
cited by other articles in ScienceCentral.
Abstract
Assessing antigen concentration of vaccine is essential step in determining the quality of the vaccine prior to vaccination. After vaccination, vaccine-induced antibody titer should also be measured to verify the vaccine efficacy. Since conventional assay used for vaccine concentrations and induced Ab-titers is antibody-based enzyme-linked immunosorbent assay, the assay inevitably brings drawbacks of antibody such as high cost for production, limited stability, and inconsistent quality between lot-to-lots. Aptamer is single-stranded nucleic acid having three-dimensional structure and has features overcoming limitations of antibody. This review will briefly introduce the features of aptamer and potential of aptamer-based system for evaluation of vaccine efficacy.
Keywords: Aptamers, Vaccines, Enzyme-linked immunosorbent assay, Antigens, Antibodies
Vaccination is a cost-effective strategy preventing a wide range of infectious diseases. To accomplish successful outcome from vaccination, exact amount of vaccine should be injected. Low amount of vaccine cannot induce enough antibodies (Ab) to prevent from infectious agents. High amount of vaccine can potentially cause organ damage due to ingredient adjuvants such as aluminum [
1]. For evaluation of vaccine efficacy, vaccine-induced Ab titers should be monitored. If Ab titer is below acceptable level or is not generated, the vaccine regimen should be adjusted. Therefore, assessment of vaccine concentration and the Ab titer are mandatory procedure before and after vaccination, respectively.
Enzyme-linked immunosorbent assay (ELISA) is a widely used method for assessment of vaccine concentration and Ab titer. Ab is a component of the assay due to its specific binding ability to the target. Production process of Ab is a well-established process. In addition, Ab-based ELISA is a relatively easy assay. Ab, however, has several drawbacks in terms of production cost and long period for generation of new Ab pairs [
2]. The limited stability and batch-dependent inconsistency of Ab are other concerns in long-term storage and quality of ELISA products to measure vaccine efficacy. In this point of view, replacing Ab with an alternative having features overcoming drawbacks of Ab in the system for evaluation of vaccine efficacy would be desirable and aptamer well meets the requirements. Later parts of this review will provide features of aptamer, aptamer-based assay, and potential use in assessment system for vaccine efficacy.
Aptamer is typically a single-stranded DNA/RNA having three-dimensional structure [
34]. The structure provides binding ability of aptamer and its binding affinity is comparable to Ab (nanomolar to picomolar range) (
Fig. 1A) [
356]. In some cases, aptamer can distinguish a single amino acid from mutated proteins [
7]. The selection process of aptamer to a specific target, called systematic evolution of ligands by exponential enrichment (SELEX), is required for approximately eight weeks for 15 rounds of SELEX screening whereas Ab selection is usually needed for 4-8 months (
Table 1,
Fig. 1B, C). The selected aptamer can be produced
in vitro by chemical synthesis with constant quality and the automated process can reduce the cost of aptamer production. Aptamer is also relatively stable in shelf because it easily returns to its initial form after a temperature change. In addition, aptamers can be easily modified to apt in various detection systems.
Since the merits of the aptamers described above are sufficient as an alternative to Ab used in detection systems, various approaches have been taken to improve conventional detection systems including ELISA. A conventional sandwich ELISA using Ab is conducted initially by coating a capture Ab on the surface of 96-well plate followed by adding a sample, and sequentially carrying out a detection Ab and a substrate bound to horseradish peroxidase. The aptamer-based ELISA process is similar as Ab-based ELISA, but modification is required for capture aptamer to immobilize the aptamer on the surface of assay plate. A representative method is using amine-modification at the end of the aptamer [
89]. Using amine-binding 96-well plate, amine-containing aptamers can be easily immobilized on the surface of the assay plate (
Fig. 2). As a modified method, the sequential immobilizing process is utilized with nucleotides having amine-moiety and complementary sequences to aptamer [
10]. By coating the nucleotides, aptamer can be immobilized on the surface of the well through sequence-specific binding between aptamer and the nucleotide. Although this immobilization process has additional step, the nucleotide can provide a space between capture aptamer and surface of assay plate. Considering that immobilized aptamer on the surface of assay plate may not have binding ability as that of free aptamer due to potential structural hindrance, the gap between the surface of assay plate and aptamer would return the binding ability of immobilized aptamer up to the same (or similar) level of free aptamer. Thus, optimizing the length of complementary nucleotides can be a critical point in designing an aptamer-based ELISA.
Using aptamer-based ELISA, several types of target proteins were detected as sensitive as Ab-based ELISA. Initial study using aptamer-based ELISA was performed to detect thrombin [
11]. The detection limit using aptamer-based ELISA was approximately 0.4 µg/mL which is 1,000-fold lower detection sensitivity than that of commercially available Ab-based ELISA. However, a recently published paper showed that detection sensitivity of aptamer-based ELISA with modification is comparable to that of Ab-based ELISA in hepato-cellular carcinoma-specific marker, lipocalin-2, suggesting that aptamer-based detection method is sufficiently sensitive for the assessment of vaccine efficacy [
12]. Recently, attempts have been made to replace Ab with an aptamer for evaluation of vaccine efficacy [
13]. In the study, carrier protein CRM
197-conjugated polysaccharide vaccine was used as a model vaccine and the concentration of the vaccine was measured by using Ab- and aptamer-based ELISA, respectively. In direct ELISA system, the aptamer system showed higher a signal-to-noise ratio compared to Ab system. However, detection sensitivity was higher in aptamer system than that of Ab system (12.6±0.5 ng/mL in aptamer-ELISA and 68-115 ng/mL in Ab-ELISA, respectively). In sandwich type assay, aptamer-based ELISA showed lower signal-to-noise ratio compared to direct-type aptamer based ELISA. However, sandwich-type aptamer-based ELISA displayed lower detection sensitivity than that by Ab-based sandwich ELISA, indicating that modification improving detection sensitivity should be followed in aptamer-based ELISA to measure concentration of vaccine and to use as a system for assessment of vaccine efficacy.
Aptamer is an attractive molecule with potentials to replace Ab in detection systems for the assessment of vaccine concentration and vaccine-induced Ab titer. As described in published papers as above, however, recent aptamer-based ELISA could not meet the standard requirements in terms of detection sensitivity, implying that further sophisticated optimization of the assay should be conducted to improve the assay using aptamer. Recently, several different types of assay systems using aptamer are being investigated and some of those systems provided comparable or even more detection sensitivity for target molecules than that of Ab-based assay [
14]. As a more advanced assay system, chip-based assay using aptamer will reduce assay time to within 5 to 10 minutes. The aptamer-based platform can provide a highly-reliable, sensitive, and speedy method to detect and measure vaccine efficacy.
Figures and Tables
 | Fig. 1Schematic cartoon for antibody (Ab) and aptamer and process of Ab and aptamer generation. (A) Structure of Ab and aptamer. (B) The process of Ab generation involves the following steps, such as mice vaccination, hybridoma generation and purification from culture supernatant of the hybridoma. (C) Aptamer is selected by systematic evolution of ligands by exponential enrichment process and amplification step in vitro.
|
 | Fig. 2Immobilization process of aptamer on assay plate. Immobilization of aptamer on surface of assay plate using amine-moiety at the aptamer (A) or amine-moiety-containing nucleotide with complementary sequences to aptamer (B).
|
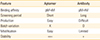
Table 1
Features of aptamer and antibody
|
Feature |
Aptamer |
Antibody |
|
Binding affinity |
pM-nM |
pM-nM |
|
Screening period |
Short |
Long |
|
Production |
Easy |
Difficult |
|
Batch variation |
X |
O |
|
Modification |
Easy |
Limited |
|
Stability |
+++ |
+ |






 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download