Abstract
Purpose
The aim of this study was to evaluate hidden allergens of acute idiopathic urticaria (AIU) in childhood by using the component-resolved diagnostics (CRD).
Methods
We applied CRD using pathogenesis-related protein family number 10 (PR-10) and nonspecific lipid transfer proteins (nsLTP).
Results
Twenty-two of the 74 AIU children (29.7%) were found to be positive on CRD. Ten children were positive to nMal d 1 for apple (value range, 1.10–40.59), 6 to rConr a 1 for hazelnut (1.53–11.97), 4 to rPru p 1 for peach (1.32–11.83). 6 to rAra h 8 for peanut (1.20–8.12), 6 to nAct d 8 for kiwi (0.85–3.32), 4 to rBet v 1 for birch (2.49–54.28), and 3 to rAln g 1 for alder (2.32–5.74). Six children were positive to nPru p 3 for peach (1.45–18.77), 4 to rCor a 8 for hazelnut (2.56–9.19), 2 to nArt v 3 for mugwort (3.40–7.42), and 3 to rBet v2 to profilin of birch (2.56–17.46). Ten children with AIU were positive to multiple component proteins. For hazelnut, 5 children were positive to PR-10 (rConr a 1) and nsLTP (rConr a 1). For peach, 3 children were positive to PR-10 (rPru p 1) and nsLTP (nPru p 3).
Figures and Tables
Table 1
The results of 22 children with component-resolved diagnostics positive in 74 acute idiopathic urticarias

Table 2
The positive results of PR-10 in children with acute idiopathic urticaria
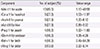
Table 3
The positive results of nsLTP and profilin in children with acute idiopathic urticaria

Table 4
Multiple positive results of PR-10, profilin, and nsLTP in children with acute idiopathic urticaria

| Source | Component | No. of subject (%) |
|---|---|---|
| Hazelnut | PR-10 (rCor a 1) and nsLTP (rCor a 1) | 4 (18.2) |
| Peach | PR-10 (rPru p 1) and nsLTP (nPru p 3) | 3 (13.6) |
| Birch | PR-10 (rBet v 1) and profilin (rBet v 2) | 2 (9.1) |
References
1. Tuncel T, Uysal P, Arikan-Ayyildiz Z, Firinci F, Karaman O, Uzuner N. Pediatricians' approach to children with acute urticaria. Minerva Pediatr. 2016; 68:96–102.
2. Carr TF, Saltoun CA. Chapter 21: urticaria and angioedema. Allergy Asthma Proc. 2012; 33:Suppl 1. S70–S72.

4. Valenta R, Duchene M, Vrtala S, Birkner T, Ebner C, Hirschwehr R, et al. Recombinant allergens for immunoblot diagnosis of tree-pollen allergy. J Allergy Clin Immunol. 1991; 88:889–894.

5. Thomas WR, Stewart GA, Simpson RJ, Chua KY, Plozza TM, Dilworth RJ, et al. Cloning and expression of DNA coding for the major house dust mite allergen Der p 1 in Escherichia coli. Int Arch Allergy Appl Immunol. 1988; 85:127–129.

6. Valenta R, Vrtala S, Ebner C, Kraft D, Scheiner O. Diagnosis of grass pollen allergy with recombinant timothy grass (Phleum pratense) pollen allergens. Int Arch Allergy Immunol. 1992; 97:287–294.

7. Valenta R, Kraft D. Recombinant allergen molecules: tools to study effector cell activation. Immunol Rev. 2001; 179:119–127.

8. Canonica GW, Ansotegui IJ, Pawankar R, Schmid-Grendelmeier P, van Hage M, Baena-Cagnani CE, et al. A WAO - ARIA - GA2LEN consensus document on molecular-based allergy diagnostics. World Allergy Organ J. 2013; 6:17.

10. Valenta R, Lidholm J, Niederberger V, Hayek B, Kraft D, Grönlund H. The recombinant allergen-based concept of component-resolved diagnostics and immunotherapy (CRD and CRIT). Clin Exp Allergy. 1999; 29:896–904.

11. Wang J, Lin J, Bardina L, Goldis M, Nowak-Wegrzyn A, Shreffler WG, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol. 2010; 125:695–702. 702.e1–702.e6.

12. Melioli G, Compalati E, Bonini S, Canonica GW. The added value of allergen microarray technique to the management of poly-sensitized allergic patients. Curr Opin Allergy Clin Immunol. 2012; 12:434–439.

13. Ruiz-García M, García Del Potro M, Fernández-Nieto M, Barber D, Jimeno-Nogales L, Sastre J. Profilin: a relevant aeroallergen? J Allergy Clin Immunol. 2011; 128:416–418.
14. Hauser M, Roulias A, Ferreira F, Egger M. Panallergens and their impact on the allergic patient. Allergy Asthma Clin Immunol. 2010; 6:1.

15. Sastre J, Landivar ME, Ruiz-García M, Andregnette-Rosigno MV, Mahillo I. How molecular diagnosis can change allergen-specific immunotherapy prescription in a complex pollen area. Allergy. 2012; 67:709–711.

16. Tripodi S, Frediani T, Lucarelli S, Macrì F, Pingitore G, Di Rienzo Businco A, et al. Molecular profiles of IgE to Phleum pratense in children with grass pollen allergy: implications for specific immunotherapy. J Allergy Clin Immunol. 2012; 129:834–839.e8.

17. Nicolaou N, Custovic A. Molecular diagnosis of peanut and legume allergy. Curr Opin Allergy Clin Immunol. 2011; 11:222–228.

18. Shreffler WG. Microarrayed recombinant allergens for diagnostic testing. J Allergy Clin Immunol. 2011; 127:843–849.

19. Borres MP, Ebisawa M, Eigenmann PA. Use of allergen components begins a new era in pediatric allergology. Pediatr Allergy Immunol. 2011; 22:454–461.

20. Eigenmann PA. Component-resolved diagnosis in food allergy, are micro-array assays helpful to the clinician? Allergy. 2008; 63:1519–1520.

21. Mittermann I, Zidarn M, Silar M, Markovic-Housley Z, Aberer W, Korosec P, et al. Recombinant allergen-based IgE testing to distinguish bee and wasp allergy. J Allergy Clin Immunol. 2010; 125:1300–1307.e3.

22. Müller U, Schmid-Grendelmeier P, Hausmann O, Helbling A. IgE to recombinant allergens Api m 1, Ves v 1, and Ves v 5 distinguish double sensitization from crossreaction in venom allergy. Allergy. 2012; 67:1069–1073.

23. Walsh J, O'Flynn N. Diagnosis and assessment of food allergy in children and young people in primary care and community settings: NICE clinical guideline. Br J Gen Pract. 2011; 61:473–475.

24. Vereda A, van Hage M, Ahlstedt S, Ibañez MD, Cuesta-Herranz J, van Odijk J, et al. Peanut allergy: clinical and immunologic differences among patients from 3 different geographic regions. J Allergy Clin Immunol. 2011; 127:603–607.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download