Abstract
During the second half of the 2000s, the significant impact of human microbiome on human diseases and health conditions was found. Since the Human Microbiome Project, many microbiome studies have been reported in domestic and international references. Gastrointestinal tract microbiome has been most investigated so far, and the association with illness has been demonstrated in many diseases. Recently, the range of study was extended to multiple human organs, such as the respiratory tract, skin, and urogenital tract. Given the scale and speed of research and development in recent years, the role of microbiome in many diseases would be established before long. In this review, we aimed to summarize the current status of microbiome studies in Korea and foreign countries with an emphasis on respiratory tract microbiome. The main concept and analytical methods for microbiome research, associations of microbiome and diseases, and research projects on Korean microbiome are reviewed.
Figures and Tables
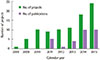
 | Fig. 1Number of the publications by National Institutes of Health (NIH) Human Microbiome Project (HMP), USA. Distribution of the publications depending on calendar year (A), and depending on associated organs or diseases (B). Data from the NIH HMP website (http://hmpdacc.org). |
Table 1
Major international microbiome projects

References
1. Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003; 300:286–290.

2. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007; 449:804–810.

4. Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001; 9:206–208.

5. Lederberg J, McCray AT. 'Ome sweet' omics: a genealogical treasury of words. Scientist. 2001; 15:8–10.
6. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012; 70:Suppl 1. S38–S44.

7. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977; 31:107–133.

8. Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012; 13:151–170.

9. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006; 7:688–693.
10. Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985; 39:321–346.

11. Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990; 345:63–65.

13. Torsvik V, Goksøyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990; 56:782–787.

14. NIH HMP Working Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome Res. 2009; 19:2317–2323.

15. Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998; 5:R245–R249.

16. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7:335–336.

17. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009; 75:7537–7541.

18. Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014; 42(Database issue):D633–D642.

19. Abarenkov K, Tedersoo L, Nilsson RH, Vellak K, Saar I, Veldre V, et al. PlutoF: a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol Bioinform. 2010; 6:189–196.
20. Bocci V. The neglected organ: bacterial flora has a crucial immunostimulatory role. Perspect Biol Med. 1992; 35:251–260.

21. Wold AE. The hygiene hypothesis revised: is the rising frequency of allergy due to changes in the intestinal flora? Allergy. 1998; 53:46 Suppl. 20–25.

22. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004; 101:15718–15723.

23. Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest. 2014; 124:4173–4181.

24. Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012; 52:37–56.

25. Saad R, Rizkallah MR, Aziz RK. Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012; 4:16.

26. Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008; 363:1–25.

27. Miller AW, Dearing D. The metabolic and ecological interactions of oxalate-degrading bacteria in the Mammalian gut. Pathogens. 2013; 2:636–652.

28. Siva S, Barrack ER, Reddy GP, Thamilselvan V, Thamilselvan S, Menon M, et al. A critical analysis of the role of gut Oxalobacter formigenes in oxalate stone disease. BJU Int. 2009; 103:18–21.

29. Jones ML, Tomaro-Duchesneau C, Prakash S. The gut microbiome, probiotics, bile acids axis, and human health. Trends Microbiol. 2014; 22:306–308.

30. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014; 30:332–338.

31. Sagar NM, Cree IA, Covington JA, Arasaradnam RP. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterol Res Pract. 2015; 2015:398585.

32. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009; 9:313–323.

33. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012; 336:1268–1273.

34. Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A. 2012; 109:8334–8339.

35. Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015; 17:565–576.

36. Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011; 108:3047–3052.

37. Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011; 105:755–764.

38. Steenbergen L, Sellaro R, van Hemert S, Bosch JA, Colzato LS. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain Behav Immun. 2015; 48:258–264.

39. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011; 474:327–336.

40. Newburg DS, Walker WA. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res. 2007; 61:2–8.

41. Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, et al. Diarrhea in young children from low-income countries leads to largescale alterations in intestinal microbiota composition. Genome Biol. 2014; 15:R76.

42. Youmans BP, Ajami NJ, Jiang ZD, Campbell F, Wadsworth WD, Petrosino JF, et al. Characterization of the human gut microbiome during travelers' diarrhea. Gut Microbes. 2015; 6:110–119.

43. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180.

44. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023.
45. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008; 3:213–223.

46. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457:480–484.

47. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–1031.

48. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013; 110:9066–9071.

49. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63:727–735.

50. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490:55–60.

51. Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiomehost interactions in inflammatory bowel diseases. Cell Host Microbe. 2015; 17:577–591.

52. Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003; 3:521–533.

53. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010; 139:1844–1854.e1.

54. Bhattacharjee S, Lukiw WJ. Alzheimer's disease and the microbiome. Front Cell Neurosci. 2013; 7:153.

56. Miklossy J. Alzheimer's disease: a neurospirochetosis. Analysis of the evidence following Koch's and Hill's criteria. J Neuroinflammation. 2011; 8:90.

57. Shoemark DK, Allen SJ. The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer's disease. J Alzheimers Dis. 2015; 43:725–738.

58. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013; 19:576–585.

59. Yi H, Yong D, Lee K, Cho YJ, Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infect Dis. 2014; 14:583.

60. Park H, Shin JW, Park SG, Kim W. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. PLoS One. 2014; 9:e109710.

61. Choi EB, Hong SW, Kim DK, Jeon SG, Kim KR, Cho SH, et al. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy. 2014; 69:517–526.

62. Jung WH, Croll D, Cho JH, Kim YR, Lee YW. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses. 2015; 58:167–172.

63. Griffin JL, Wang X, Stanley E. Does our gut microbiome predict cardiovascular risk? A review of the evidence from metabolomics. Circ Cardiovasc Genet. 2015; 8:187–191.

64. Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014; 124:4204–4211.

65. DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015; 112:11060–11065.

66. Metcalf JL, Xu ZZ, Weiss S, Lax S, Van Treuren W, Hyde ER, et al. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science. 2016; 351:158–162.

67. Emanuelsson F, Claesson BE, Ljungström L, Tvede M, Ung KA. Faecal microbiota transplantation and bacteriotherapy for recurrent Clostridium difficile infection: a retrospective evaluation of 31 patients. Scand J Infect Dis. 2014; 46:89–97.

68. Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011; 53:994–1002.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download