Dear Editor,
Retinitis pigmentosa (RP, OMIM 268000) is the most common disorder among a group of inherited progressive retinal dystrophies (RD) and is characterized by retinal pigment deposits visible on fundus examination and primary loss of rod photoreceptor cells followed by secondary loss of cone photoreceptors [1]. RP is a clinically and genetically heterogeneous disorder with a prevalence of 1 in 4,000 individuals [1234]. More than 70 genes are known to be involved in RP (Table 1, https://sph.uth.edu/RETNET), and hence, the identification of causative variations in known candidate genes is challenging. Recently, diagnostic exome sequencing (DES) has been introduced to analyze the exons and flanking intronic regions of genes with clinical relevance in heterogeneous diseases such as RP [5].
A 35-year-old woman with symptoms of night blindness and difficulty seeing well in dim light presented for an eye examination. She had no family history of retinal disease. Her best-corrected visual acuity was 20/20 in the right eye and 20/20 in the left eye. The refractive error was −2.50 D in the right eye and −2.75 D in the left eye. Dilated fundus examination showed attenuated retinal blood vessels and perivascular bone-spicule deposits with diffuse depigmented dots in the periphery of both eyes. Optic disc pallor was not observed (Fig. 1A).
Full-field electroretinography (Roland, RETI-port/scan21, Brandenburg, Germany) revealed near absence of photopic and scotopic responses in both eyes (Fig. 1B). Visual field testing (Humphrey 740i central 30-2 program, Zeiss, Jena, Germany) demonstrated a constricted visual field in both eyes (Fig. 1C).
Spectral domain optical coherence tomography (SD OCT; Optovue RTVue XR Avanti, Optovue Inc., CA, USA) and fundus autofluorescence imaging (FAF) (Spectralis OCT, Heidelberg Engineering, Heidelberg, Germany) showed areas of photoreceptor loss outside of the central macula, corresponding to disappearance of the junction between the inner segment and outer segment (IS/OS) line and hypoautofluorescence, respectively (Fig. 1D and E). The retinal layers in the central macula were relatively preserved. Significantly reduced choroidal thickness was also found in both eyes. En face structural and angiographic imaging with SD OCT of both macular lesions revealed choriocapillaris in the peripheral region in greater detail compared to the central region, with an intact outer retina due to increased light penetration in the atrophic peripheral macula (Fig. 1F).
The methodology of DES was the same as that in a previously published case by Jang et al [6].Genomic DNA was enriched by using the TruSight One Sequencing Panel (Illumina, Inc., San Diego, CA, USA), which includes 25,396 probes targeting a 12-Mb region spanning 4,813 genes, and sequenced on the Illumina MiSeq platform. Raw sequence reads were processed and aligned to the UCSC hg19 human reference sequence with the Burrows-Wheeler Aligner (BWA, version 0.7.12). Duplicate reads were removed with Picard and local alignment optimization was performed with the Genome Analysis Tool Kit (GATK, version 3.5). The single nucleotide polymorphism (SNP) and short indel candidates were identi-fied and these variants were annotated by ANNOVAR to filter SNPs reported in the SNP database (dbSNP, build 129) and the 1,000 Genomes Project (http://1000genomes.org). Four types of online predictive software, including Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/) [7], and Polymorphism Phenotyping v2 (PolyPhen-2, v.2.2.2; http://genetics.bwh.harvard.edu/pph2/) [8] were applied to predict the impact of the variants. Nucleotide conservation between mammalian species was evaluated by using the Evolutionary Annotation Database (Evola version 7.5, http://www.h-invitational.jp/evola/) [9]. A mean coverage of 113.25× was achieved, and 95.7% of targeted bases were read >20 times by exome capture and sequencing. A total of 8,006 variants including 7,610 SNVs were identified, and after exclusion of nongenic intronic variants and synonymous exonic variants, 352 variants remained. Among these, common polymorphisms listed in dbSNP129 with a minor allele frequency (MAF) >0.05 were excluded.
DES revealed two known pathogenic variants in the EYS (eyes shut homolog) gene (reference sequence, NM_001142800.1)—c.6563T>C (p.Ile2188Thr) and c.9059T>C (p.Ile3020Thr)—which were validated by Sanger sequencing. Further analysis of the family confirmed that c.6563T>C (p.Ile2188Thr) was inherited from the mother (Fig. 2). The origin of c.9059T>C could not be elucidated because the proband's father was deceased. The c.6563T>C (p.Ile2188Thr) variant was previously reported by Arai et al [4], and the c.9059T>C (p.Ile3020Thr) variant was reported by Di et al [10]. On the basis of the ACMG/AMP 2015 criteria, these two variants were classified as likely pathogenic variants.
The EYS gene located at the RP25 locus on chromosome 6q12 was first discovered to be commonly mutated in autosomal recessive RP by Mai et al [2]. With 43 exons covering 2.0 Mb, EYS is the largest gene shown to be expressed in the human eye and the fifth largest overall in the human genome, encoding a large protein of 3,165 amino acids with multiple domains, including 28 epidermal growth factor (EGF) or EGF-like domains and five laminin G domains [11]. The two variants found in our patient were in the second and fifth laminin G domains, respectively, which could be lost due to the resulting conformational changes [12].
These observations suggest a possible role of the EYS protein in human retinal integrity, but the exact mechanism underlying degeneration of photoreceptor cells due to pathogenic variants in the EYS gene requires further investigation. In previous studies by Iwanami et al [3], the variant type was related to the severity of RP symptoms. More studies on the correlation between the RP genotype and phenotype would enable better diagnosis and improve prediction of the incidence of RP. Autosomal recessive RP accounts for up to 17.3% of cases of RP, and sporadic RP is the most common (61.9%) form in Koreans [13]. In summary, we report the first case of RP related to missense variants of the EYS gene in Korea. This report will contribute to a better understanding of the genetic background of Korean RP patients.
Figures and Tables
Figure 1
A 35-year-old woman with retinitis pigmentosa. Her best-corrected visual acuity was 20/20 in the right eye and 20/20 in the left eye. (A) Color fundus photographs of both eyes show mottling and granularity at the level of the retinal pigment epithelium (RPE), perivascular bone spicule-shaped pigment deposits in the peripheral retina, and attenuated retinal vessels. The picture on the left is the right eye, and that on the right is the left eye. (B) Full field electroretinography (ERG) reveals near absence of both photopic and scotopic responses of both eyes. The a-wave is derived from the cones and rods of the outer photoreceptor layers. The bwave is derived from the inner retina, predominantly Müller glia and ON-bipolar cells. P2 is the bipolar cell-derived component of the rod-isolated b-wave, and P3 is subtracted from the series of rod responses. The left graph shows the right eye, and the right graph shows the left eye. (C) Visual field testing demonstrates marked peripheral loss with a small residual central visual field in both eyes. The picture on the left is the left eye, and that on the right is the right eye. (D) The lack of signal on fundus autofluorescence imaging (FAF) correlates well with the areas of RPE atrophy. FAF demonstrated a perifoveal ring of increased autofluorescence within the macula with thread-like retinal vessels. The picture on the left is the right eye, and that on the right is the left eye. (E) Spectral domain optical coherence tomography (SD-OCT) shows decreased retinal thickness, particularly in the outer layer: decreased thickness of the outer nuclear layer and loss of the external limiting membrane and the junction between inner segments and outer segments (IS/OS) line (between yellow arrows). The upper picture is the right eye, and the lower is the left eye. (F) En face structural and angiographic imaging with SD-OCT (inset shows a scan plane taken just below the Bruch's membrane) reveals choriocapillaris in the peripheral region in greater detail compared to the central region, with intact outer retina due to increased light penetration in the atrophic peripheral macula. The upper picture is the right eye, and the lower is the left eye.
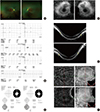
Figure 2
(A) IGV browser visualization of the exome sequencing results of partial genomic DNA sequence of the EYS gene of the patient shows two heterozygous variants. (Upper, c.6563T>C, Lower, c.9059T>C) (B) Validation of EYS variants by Sanger sequencing. The patient had two non-synonymous substitutions—c.[6563T>C];[9059T>C] (arrow)—in EYS. The patient's mother possessed c.6563T>C, but not c.9059T>C.

Table 1
Genes causing retinitis pigmentosa (https://sph.uth.edu/RETNET/)

References
2. Abd El-Aziz MM, Barragan I, O'Driscoll CA, Goodstadt L, Prigmore E, Borrego S, et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet. 2008; 40:1285–1287.

3. Iwanami M, Oshikawa M, Nishida T, Nakadomari S, Kato S. High prevalence of mutations in the EYS gene in Japanese patients with autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2012; 53:1033–1040.

4. Arai Y, Maeda A, Hirami Y, Ishigami C, Kosugi S, Mandai M, et al. Retinitis pigmentosa with EYS mutations is the most prevalent inherited retinal dystrophy in Japanese populations. J Ophthalmol. 2015; 2015:819760.
5. Yoon CK. Strategies for mutation discovery in retinitis pigmentosa: transition to the next generation. J Genet Med. 2013; 10:13–19.

6. Jang MA, Lee T, Lee J, Cho EH, EH CS. Identification of a novel de novo variant in the PAX3 gene in Waardenburg syndrome by diagnostic exome sequencing: the first molecular diagnosis in Korea. Ann Lab Med. 2015; 35:362–365.

7. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009; 4:1073–1081.

8. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–249.

9. Matsuya A, Sakate R, Kawahara Y, Koyanagi KO, Sato Y, Fujii Y, et al. Evola: Ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008; 36:D787–D792.

10. Di Y, Huang L, Sundaresan P, Li S, Kim R, Ballav Saikia B, et al. Whole-exome sequencing analysis identifies mutations in the EYS gene in retinitis pigmentosa in the Indian population. Sci Rep. 2016; 6:19432.

11. Gurudev N, Yuan M, Knust E. chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells. Biol Open. 2014; 3:332–341.

12. Khan MI, Collin RW, Arimadyo K, Micheal S, Azam M, Qureshi N, et al. Missense mutations at homologous positions in the fourth and fifth laminin A G-like domains of eyes shut homolog cause autosomal recessive retinitis pigmentosa. Mol Vis. 2010; 16:2753–2759.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download