Abstract
We report a patient with massive eosinophilia and a complex karyotype that was initially misdiagnosed as chronic eosinophilic leukemia (CEL), but later diagnosed as anaplastic large cell lymphoma (ALCL) masked by massive eosinophilia. The complex karyotype observed at initial diagnosis remained unchanged later, after the evidence of bone marrow involvement of ALCL was obtained. At diagnosis, genetic aberrations corresponding to metaphase cytogenetics were not identified by interphase fluorescence in situ hybridization, although abnormal results were noted at follow-up. Together, these observations indicate that the complex karyotype at initial work-up has been derived from a low proportion of lymphoma cells with high mitotic ability that were not identified by microscopy, rather than from massive eosinophils. These findings suggest that our patient had ALCL with secondary eosinophilia rather than CEL since initial diagnosis.
Go to : 
REFERENCES
1.Gotlib J. World Health Organization-defned eosinophilic disorders: 2015 update on diagnosis, risk stratifcation, and management. Am J Hematol. 2015. 90:1077–89.
2.Ogata M., Ogata Y., Kohno K., Uno N., Ohno E., Ohtsuka E, et al. Eosinophilia associated with adult T-cell leukemia: role of interleukin 5 and granulocyte-macrophage colony-stimulating factor. Am J Hematol. 1998. 59:242–5.

3.Montgomery ND., Dunphy CH., Mooberry M., Laramore A., Foster MC., Park SI, et al. Diagnostic complexities of eosinophilia. Arch Pathol Lab Med. 2013. 137:259–69.

5.Desenne JJ., Acquatella G., Stern R., Muller A., Sanchez M., Somoza R. Blood eosinophilia in Hodgkin's disease. A follow-up of 25 cases in Venezuela. Cancer. 1992. 69:1248–53.

6.Utsunomiya A., Ishida T., Inagaki A., Ishii T., Yano H., Komastsu H, et al. Clinical signifcance of a blood eosinophilia in adult T-cell leukemia/lymphoma: a blood eosinophilia is a signifcant unfavorable prognostic factor. Leuk Res. 2007. 31:915–20.
7.McKelvie PA., Oon S., Romas E., Nandurkar H., Tam CS. A case of systemic anaplastic lymphoma kinase-negative anaplastic large cell lymphoma associated with hypereosinophilia, granulomatous myositis and vasculitis. Leuk Lymphoma. 2012. 53:2279–82.

8.Orofno N., Guidotti F., Cattaneo D., Sciume M., Gianelli U., Cortelezzi A, et al. Marked eosinophilia as initial presentation of breast implant-associated anaplastic large cell lymphoma. Leuk Lymphoma. 2016. 57:2712–5.
9.Powers ML., Watson BW., Frater JL., Kreisel F., Hassan A. Peripheral eosinophilia camoufaging anaplastic large cell lymphoma. Int J Surg Pathol. 2011. 19:405–8.
10.Gonsalves WI., He R., Pardanani A., Gupta V., Smeltzer JP., Hanson CA, et al. Chronic eosinophilic leukemia-not otherwise specifed (NOS) in the background of a large cell lymphoma. Case Rep Hematol. 2013. 2013:458303.
11.Fan YS., Rizkalla K. Comprehensive cytogenetic analysis including multicolor spectral karyotyping and interphase fuorescence in situ hybridization in lymphoma diagnosis. a summary of 154 cases. Cancer Genet Cytogenet. 2003. 143:73–9.
13.Mereu E., Pellegrino E., Scarfo I., Inghirami G., Piva R. The heterogeneous landscape of ALK negative ALCL. Oncotarget. 2017. 8:18525–36.

Go to : 
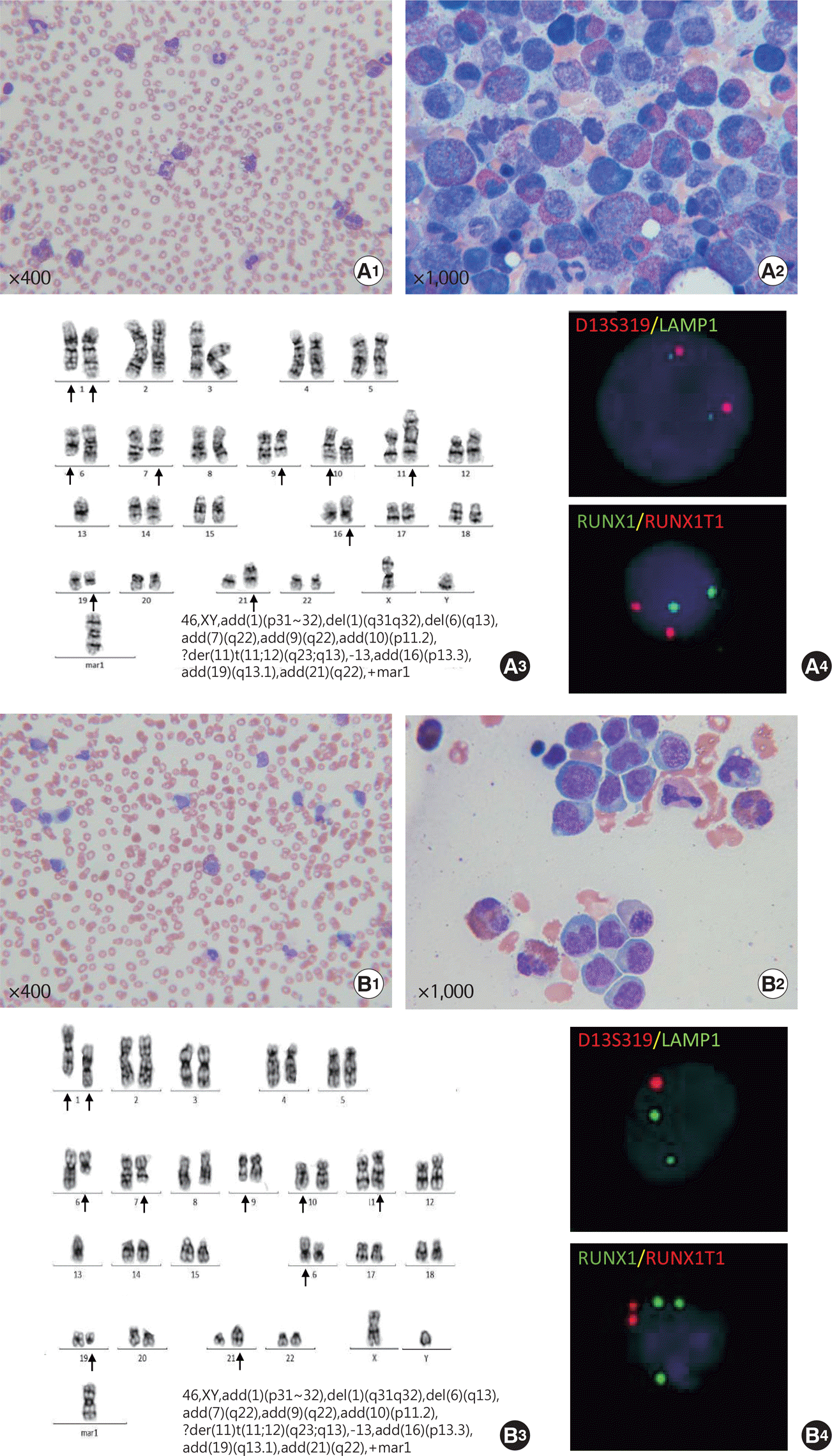 | Fig. 1.Morphology, cytogenetic studies, and interphase fluorescence in situ hybridization (iFISH) at diagnosis (A) and follow-up (B). (A1) Peripheral blood smear showing eosinophilia without abnormal lymphoid cells. (A2) BM examination showing hypercellularity with marked eosinophil predominance. No abnormal lymphoid cells were observed. (A3) G-banded karyotype result showing a complex karyotype at diagnosis. (A4) iFISH using D13S319/13q34 probes and RUNX1/RUNX1T1 probes shows normal hybridization signals. (B1) Peripheral blood smear revealing abnormal lymphoid cells with eosinophilia. (B2) BM aspiration revealing large irregular lymphoid cells with some eosinophils. (B3) G-banded karyotype revealing the same complex karyotype at follow-up as at diagnosis. (B4) iFISH using D13S319/13q34 probes shows two green signals (LAMP1) and one orange (D13S319) signal, indicating deletion of the 13q14.3 locus. iFISH using RUNX1/RUNX1T1 probes shows three green (RUNX1) and two orange (RUNX1T1) signals, demonstrating RUNX1 amplification. |
Table 1.
Laboratory characteristics at initial diagnosis and follow-up study
| At initial diagnosis | One year after initial diagnosis | |
|---|---|---|
| Peripheral blood | ||
| Hemoglobin (g/dL) | 13.9 | 8.0 |
| Platelet (×109/L) | 149 | 33 |
| WBC (×109/L) | 49.7 | 45.3 |
| Differential count | ||
| Segmented neutrophils (%) | 17 | 29 |
| Lymphocyte (%) | 5 | 7 |
| Monocyte (%) | 7 | 3 |
| Eosinophil (%) | 70 | 28 |
| Basophil (%) | 1 | 1 |
| Abnormal lymphocyte (%) | 0 | 32 |
| Absolute eosinophil count (×109/L) | 34.8 | 12.7 |
| Flow cytometry (% of lymphocyte cells) | ||
| Surface CD3+ | 61.0% | 1.1% |
| Cytoplasmic CD3+ | NT | 59.0% |
| CD3-/CD4+ | NT | 97.4% |
| CD3-/CD8+ | NT | 0.6% |
| CD3+/CD4+ | 48.0% | 0.3% |
| CD3+/CD8+ | 9.0% | 0.8% |
| CD19+ | 19.0% | 0.0% |
| CD16+/CD56+ | 7.0% | 5.9% |
| Bone marrow study | ||
| Cellularity | Hypercellular (80–90%) | Normocellular (40–50%) |
| Myeloblast (%) | 0.2 | 1 |
| Eosinophil (%) | 46.2 | 20.6 |
| Lymphocyte (%) | 4.8 | 16.8 |
| Abnormal lymphocyte (%) | not observed | 11 |
| Immunohistochemistry | ||
| CD30 | Negative | Negative |
| CD3 | Negative | Negative |
| CD20 | Negative | NT |
| CD4 | Negative | Positive in some aggregates |
| CD8 | Negative | NT |
| ALK | Negative | Negative |
| Cytogenetic/Molecular study | ||
| Karyotype | 46,XY,add(1)( p31~32),del(1)(q31q32),del(6)(q13),add(7) | 46,XY,add(1)( p31~32),del(1)(q31q32),del(6)(q13),add(7) |
| (q22),add(9)(q22),add(10)(p11.2),?der(11)t(11;12)(q23;q13), | (q22),add(9)(q22),add(10)(p11.2),?der(11)t(11;12)(q23;q13), | |
| -13,add(16)(p13.3),add(19)(q13.1),add(21) | -13,add(16)(p13.3),add(19)(q13.1),add(21) | |
| (q22),+mar[13]/46,XY[7] | (q22),+mar[7]/46,XY[13] | |
| iFISH | ||
| BCR/ABL1 rearrangement∗ | Negative (0.0%) | NT |
| FIP1L1/PDGFRA rearrangement∗ | Negative (0.0%) | NT |
| PDGFRB rearrangement∗ | Negative (0.5%) | NT |
| KMT2A (MLL) rearrangement∗ | Negative (0.5%) | Negative (0.0%) |
| 13q14.3deletion† | Negative (1.0%) | Positive (53.5%) |
| RUNX1 amplification∗ | Negative (0.0%) | Positive (47.0%) |
| 6q21 deletion∗ | NT | Positive (19.0%) |
| 7q31 amplification∗ | Negative (0.5%) | Positive (30.0%) |
Abbreviations: ALK, anaplastic lymphoma kinase; FIP1L1/PDGFRA, FIP1-like-1–platelet-derived growth factor receptor-alpha; iFISH, interphase fluorescence in situ hybridization; MLL, myeloid lymphoid leukemia, NT, not tested; PDGFRB, platelet-derived growth factor receptor-beta; RUNX1, Runt-related transcription factor.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download