Abstract
Background
Clostridium difficile is a leading causative microorganism of pseudomembranous colitis (PMC) and antibiotic-associated diarrhea. In patients who have a history of antibiotic use and diarrhea, the presence of the C. difficile toxin should be confirmed to diagnose C. difficile infection (CDI). In this study, the results of three assays for CDI, which were performed on 1,363 clinical stool samples at a tertiary hospital, were analyzed to evaluate the performance and usefulness of these assays for diagnosis of CDI.
Methods
The results of the VIDAS C. difficile Toxin A&B Immunoassay (bioMérieux SA, France), Xpert C. difficile Real-Time PCR Assay (Cepheid, USA), and ChromID C. difficile Agar (bioMérieux SA, France) culture were analyzed retrospectively. Cases were defined as CDI according to the positive Xpert assay or the positive VIDAS assay and/or culture in the presence of PMC findings after radiological imaging or endoscopic procedures.
Results
A total of 1,027 samples (75.8%) tested negative in all three assays, 101 samples (7.4%) tested positive in all three assays, and overall agreement among them was 82.7%. In this study, 291 cases (21.3%) were diagnosed as CDI. Sensitivity and specificity of the VIDAS assay were 38.8% and 99.3%, and those of ChromID culture were 71.5% and 96.5%, respectively. The Xpert assay showed good sensitivity (98.6%, 287/291), whereas the VIDAS assay and ChromID culture showed low sensitivities.
Figures and Tables
Table 1
Summary of the results of three diagnostic assays for CDI from 1,363 stool samples
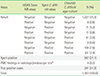
Table 2
Agreement on results in comparison with the Xpert C. difficile assay

| OPA | PPA | NPA | κ coefficient (95% CI) | P value | |
|---|---|---|---|---|---|
| % | |||||
| VIDAS | 86.6 | 39.0 | 99.3 | 0.487 (0.423–0.543) | < 0.05 |
| ChromID | 91.0 | 71.4 | 96.2 | 0.714 (0.668–0.759) | < 0.05 |
References
1. Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978; 298:531–534.

2. Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systemic review. JAMA. 2015; 313:398–408.

3. Swindells J, Brenwald N, Reading N, Oppenheim B. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol. 2010; 48:606–608.

4. Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010; 48:889–893.

5. Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, et al. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol. 2010; 48:3719–3724.

6. Knetsch CW, Bakker D, de Boer RF, Sanders I, Hofs S, Kooistra-Smid AM, et al. Comparison of real-time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection. J Clin Microbiol. 2011; 49:227–231.

7. Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difiicile/Epi, and Illuigene C. difficile assays. J Clin Microbiol. 2012; 50:1331–1335.

8. Sloan LM, Duresko BJ, Gustafson DR, Rosenblatt JE. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol. 2008; 46:1996–2001.

9. Karre T, Sloan L, Patel R, Mandrekar J, Rosenblatt J. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2011; 49:725–727.

10. Yoo J, Lee H, Park KG, Lee GD, Park YG, Park YJ. Evaluation of 3 automated real-time PCR (Xpert C. difficile assay, BD MAX Cdiff, and IMDx C. difficile for Abbott m2000 assay) for detecting Clostridium difficile toxin gene compared to toxigenic culture in stool specimens. Diagn Microbiol Infect Dis. 2015; 83:7–10.

11. Carroll KC, Loeffelholz M. Conventional versus molecular methods for the detection of Clostridium difficile. J Clin Microbiol. 2011; 49:S. 49–52.

12. Whang DH, Joo SY. Evaluation of the diagnostic performance of the Xpert Clostridium difficile assay and its comparison with the toxin A/B enzyme-linked fluorescent assay and in-house real-time PCR assay used for the detection of toxigenic C. difficile. J Clin Lab Anal. 2014; 28:124–129.

13. Yang JJ, Nam YS, Kim MJ, Cho SY, You E, Soh YS, et al. Evaluation of a chromogenic culture medium for the detection of Clostridium difficile. Yonsei Med J. 2014; 55:994–998.

14. Sharp S, Gilligan PH. A practical guidance document for the laboratory detection of toxigenic Clostridium difficile. Washington, D.C.: ASM Public and Scientific Affairs Board (PSAB) Committee on Laboratory Practices, American Society for Microbiology (ASM);2010. Updated on Sep 2010. http://www.asm.org/images/pdf/Clinical/clostridiumdifficile9-21.pdf.
15. Verhoye E, Vandecandelaere P, De Beenhouwer H, Coppens G, Cartuyvels R, Van den Abeele A, et al. A hospital-level cost-effectiveness analysis model for toxigenic Clostridium difficile detection algorithms. J Hosp Infect. 2015; 91:123–128.

16. Tenover FC, Baron EJ, Peterson LR, Persing DH. Laboratory diagnosis of Clostridium difficile infection: can molecular amplification methods move us out of uncertainty? J Mol Diagn. 2011; 13:573–582.
17. Brecher SM, Novak-Weekley SM, Nagy E. Laboratory diagnosis of Clostridium difficile infections: there is light at the end of colon. Clin Infect Dis. 2013; 57:1175–1181.

18. Riley TV, Brazier JS, Hassan H, Williams K, Phillips KD. Comparison of alcohol shock enrichment and selective enrichment for the isolation of Clostridium difficile. Epidemiol Infect. 1987; 99:355–359.

19. Clabots CR, Gerding SJ, Olson MM, Peterson LR, Gerding DN. Detection of asymptomatic Clostridium difficile carriage by an alcohol shock procedure. J Clin Microbiol. 1989; 27:2386–2387.

21. McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005; 353:2433–2441.

22. Kuijper EJ, Coignard B, Tüll P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006; 12:2–18.

23. Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg Infect Dis. 2007; 13:1417–1419.

24. Yoldaş Ö, Altındiş M, Cufalı D, Aşık G, Keşli R. A Diagnostic algorithm for the detection of Clostridium difficile-associated diarrhea. Balkan Med J. 2016; 33:80–86.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download