Abstract
Background
We evaluated a sensitive and quantitative method utilizing fragment analysis of the fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD), simultaneously measuring mutant allele burden and length, and verified the analytical performance.
Methods
The number and allelic burden of FLT3-ITD mutations was determined by fragment analysis. Serial mixtures of mutant and wild-type plasmid DNA were used to calculate the limit of detection of fragment analysis, conventional PCR, and Sanger sequencing. Specificity was evaluated using DNA samples derived from 50 normal donors. Results of fragment analysis were compared to those of conventional PCR, using 481 AML specimens.
Results
Defined mixtures were consistently and accurately identified by fragment analysis at a 5% relative concentration of mutant to wild-type, and at 10% and 20% ratios by conventional PCR and direct sequencing, respectively. No false positivity was identified. Among 481 AML specimens, 40.1% (193/481) had FLT3-ITD mutations. The mutant allele burden (1.7–94.1%; median, 28.2%) and repeated length of the mutation (14–153 bp; median, 49 bp) were variable. The concordance rate between fragment analysis and conventional PCR was 97.7% (470/481). Fragment analysis was more sensitive than conventional PCR and detected 11 additional cases: seven had mutations below 10%, three cases represented conventional PCR failure, and one case showed false negativity because of short ITD length (14 bp).
Go to : 
REFERENCES
1. Kiyoi H, Towatari M, Yokota S, Hamaguchi M, Ohno R, Saito H, et al. Internal tandem duplication of the FLT3 gene is a novel modality of elongation mutation which causes constitutive activation of the product. Leukemia. 1998; 12:1333–7.

2. Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008; 358:1909–18.

3. Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2012; 115:453–74.

4. Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, et al. Prevalence and prognostic signifcance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001; 97:89–94.
5. Estey EH. Acute myeloid leukemia: 2013 update on risk-stratifcation and management. Am J Hematol. 2013; 88:318–27.
6. O'Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, et al. Acute myeloid leukemia, version 2.2013. J Natl Compr Canc Netw. 2013; 11:1047–55.
7. Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008; 111:2776–84.

8. Meshinchi S, Stirewalt DL, Alonzo TA, Boggon TJ, Gerbing RB, Rocnik JL, et al. Structural and numerical variation of FLT3/ITD in pediatric AML. Blood. 2008; 111:4930–3.

9. Kim Y, Lee GD, Park J, Yoon JH, Kim HJ, Kim M, et al. Quantitative fragment analysis of FLT3-ITD effciently identifying poor prognostic group with high mutant allele burden or long ITD length. Blood Cancer J. 2015. e336.
10. Shires IS, Gerdener T. FLT3 ITD detection: a closer look at the options. Med Technol SA. 2011; 25:39–46.
11. Beretta C, Gaipa G, Rossi V, Bernasconi S, Spinelli O, Dell'Oro MG, et al. Development of a quantitative-PCR method for specifc FLT3/ITD monitoring in acute myeloid leukemia. Leukemia. 2004; 18:1441–4.
12. Murphy KM, Levis M, Hafez MJ, Geiger T, Cooper LC, Smith BD, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003; 5:96–102.

Go to : 
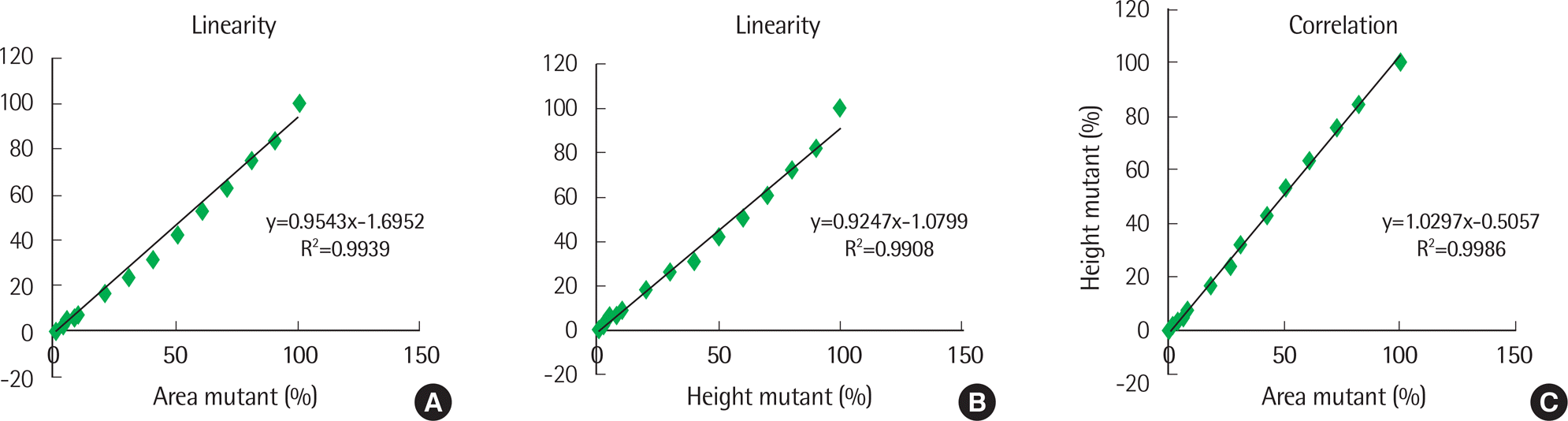 | Fig. 1.(A) Linearity of fragment analysis using standard material (66 bp-type), calculating percent ratio for area of peak. (B) Linearity of fragment analysis using standard material (66 bp-type), calculating percent ratio for height of peak. (C) Correlation for fragment analysis using methods (A) and (B). |
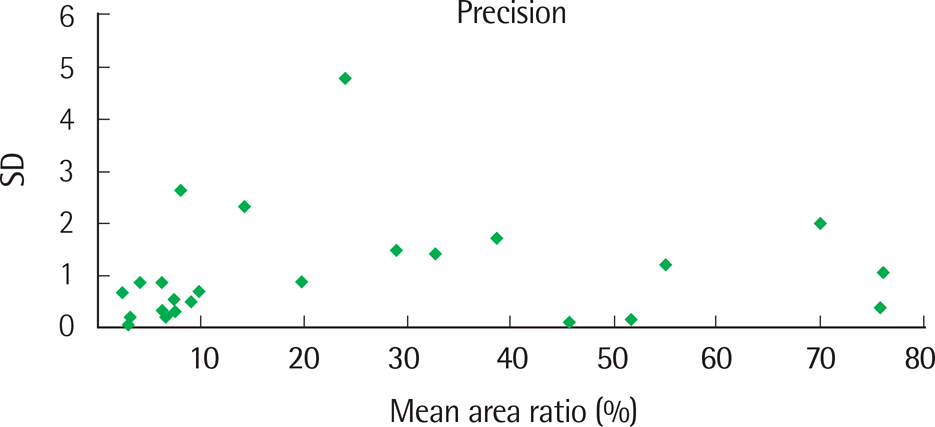 | Fig. 2.Distribution of standard deviation (SD) values of the mean area ratio by fragment analysis, using patient specimens. |
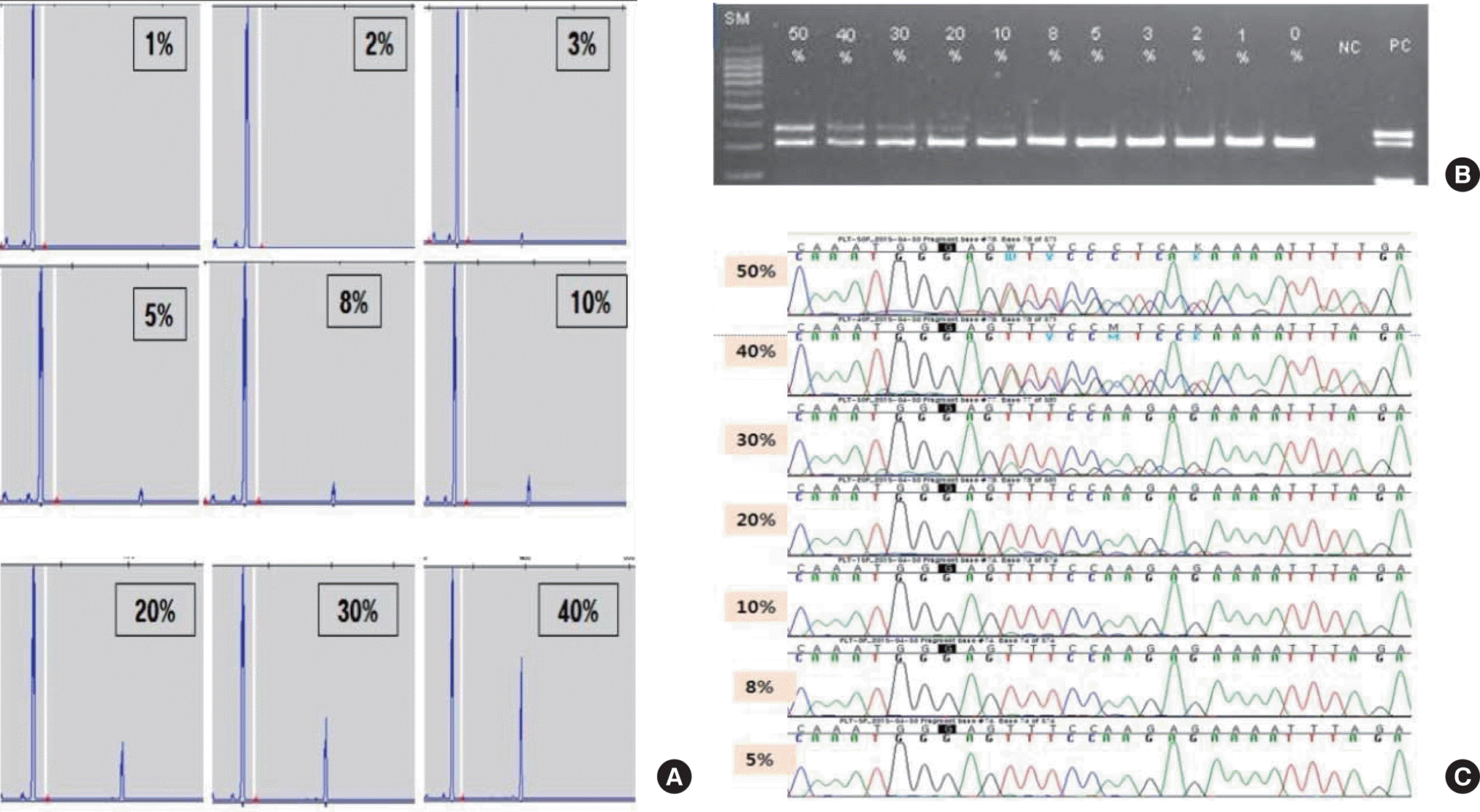 | Fig. 3.Representative examples of limit of detection for each method; the results of fragment analysis [(A) was 3%], conventional PCR [(B) was 10%], Sanger sequencing [(C) was 20%]. |
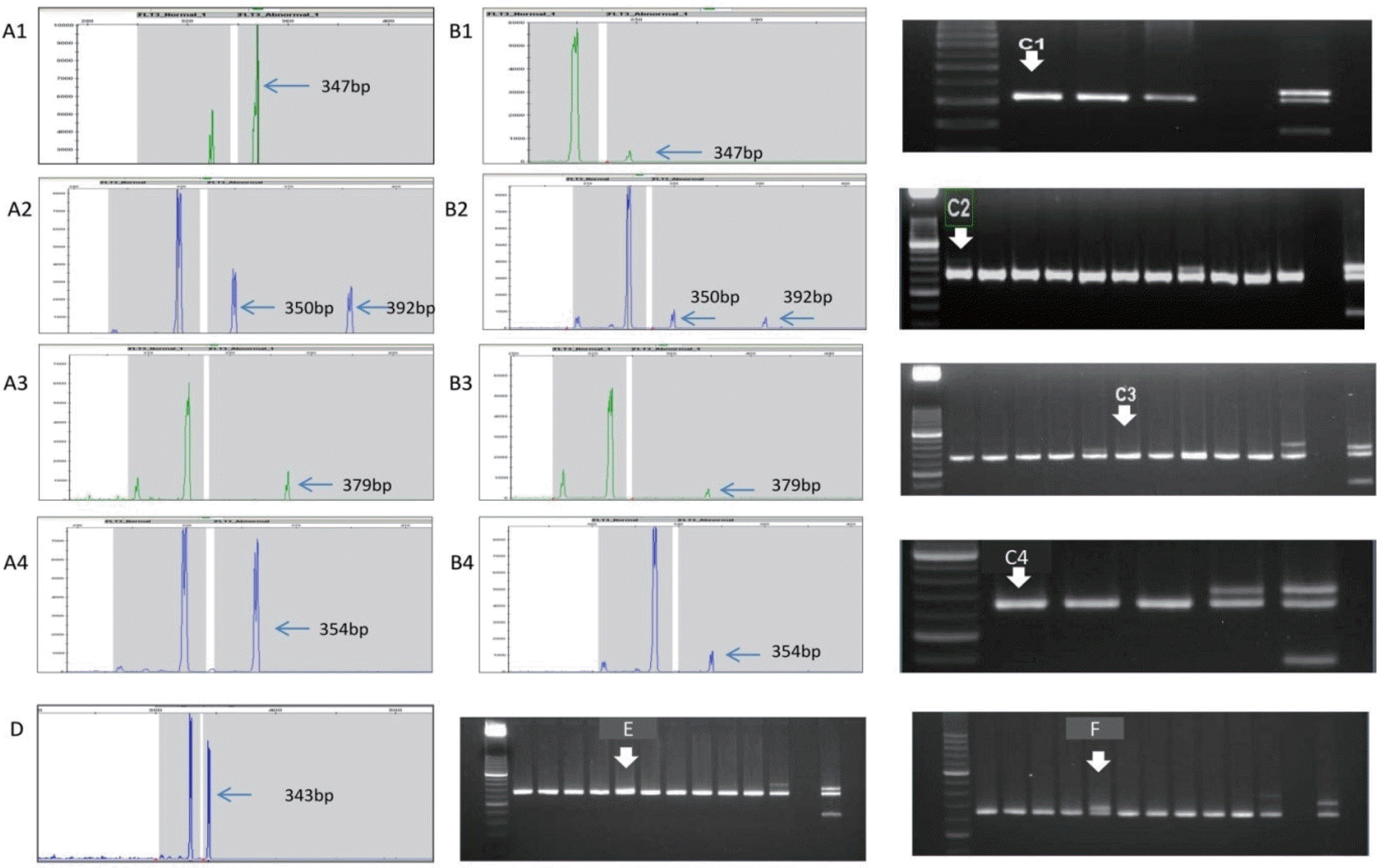 | Fig. 4.fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) analysis in acute myeloid leukemia patients: fragment analysis versus conventional PCR in discrepant case. Fig. 4. fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) analysis in acute myeloid leukemia patients: fragment analysis versus conventional PCR in discrepant case. (A) Fragment analysis for initial sample; the arrows indicating peak size are FLT3-ITDs. (B) Fragment analysis for followup sample of (A); the arrows indicating the smaller peak are the same as the peak size of (A). (C) Conventional PCR for followup of sample of (A), which used the same sample as used in (B); In the photographs, it was difficult to identify the existence of an FLT3-ITD-positive band. (D) Fragment analysis for short-length ITD. (E) For conventional PCR, this is shown as a slightly thick wild-type band when electrophoresis was performed under typical conditions. (F) To confirm the ITD band, we extended the running time of electrophoresis; in doing this, the wild-type band and ITD band could be distinguished. |
Table 1.
Distribution and length of FLT3-ITD mutations found in 193 samples by fragment analysis
| FLT3-ITD Length (bases) | Number of positive samples | Number of positive patients |
|---|---|---|
| 1–19 | 13 | 10 |
| 20–29 | 35∗† | 29 |
| 30–39 | 27∗ | 21 |
| 40–49 | 25 | 21 |
| 50–59 | 22 | 19 |
| 60–69 | 34† | 22 |
| 70–79 | 14 | 11 |
| 80–89 | 9 | 7 |
| 90–99 | 8 | 6 |
| 100–109 | 3 | 3 |
| ≥110 | 5 | 5 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download