INTRODUCTION
Sepsis is a critical illness with associated mortality ranging from 14 to 34% [
12]. Early and accurate diagnosis of sepsis is crucial for appropriate patient management. Currently, the most reliable approach to confirm sepsis is blood culture. Resin media have been recently applied to the BacT/Alert 3D System (bioMérieux Inc., Durham, NC, USA). Resin may adsorb antibiotics and other host immune factors in the bottles and therefore enhance the growth of microorganisms. Although these BacT/Alert 3D resin media have been utilized in the clinical laboratory for a few years, prospective clinical evaluation is not yet sufficient.
Blood volume is the most important parameter for successful blood culture [
13]. Although 20-30 mL of blood is recommended for each puncture, a survey revealed the mean blood volume to be only 7.7 mL per set in nine university-affiliated hospitals [
4]. Other studies also showed under-filled blood volumes for blood cultures, especially for seriously ill patients [
5]. Another study reported that many laboratories were using a 10-mL protocol for one set of blood culture [
13].
Although blood cultures should be performed before antibiotic treatment, there have been many cases of prior antibiotic use, especially for those who are admitted to intensive care units [
67]. We hypothesized that the rate of positivity (for microorganisms) would be equal for patients with or without prior antibiotic treatment due to the effect of resin. The necessity of terminal subculture has not been clearly documented for the resin bottles without a positive signal after 5 days of incubation. Early detection of microorganisms is a highlight of the automated continuous blood culture systems. Rapid detection and early reports of blood culture are crucial for treating sepsis patients and for reducing hospital stays [
189]. Therefore, detection time is an important indicator of performance in the evaluation of blood culture systems.
The aim of this study was to evaluate the clinical performance of the blood culture resin media FA Plus (aerobic) and FN Plus (anaerobic) of the BacT/Alert 3D System. Positivity rate and time to detection (TTD) were analyzed for clinically relevant microorganisms. All signal-negative bottles were subcultured to evaluate false negatives in the blood culture system. In addition, a history of prior antibiotic use was investigated to determine if this influences the effect of resin in blood cultures.
MATERIALS AND METHODS
Patients and Sample Collection
The study was conducted at the Gyeongsang National University Hospital in Korea from June to December 2014 and was approved by the IRB (GNUH 2015-03-015). Among the adult patients (≥ 18 yr of age) who visited the emergency department (ED) and were admitted to the medical and surgical intensive care units (MICU, SICU), those patients with blood culture tests based on the suspicion of sepsis were included in this study. We obtained 2,994 sets of blood cultures, 2,516 sets from the ED, 366 sets from the MICU, and 112 sets from the SICU. Well-trained and educated medical technicians collected blood samples in the ED, whereas intern physicians collected samples in the intensive care units. Two sets of blood cultures were collected for most of episodes. Chlorhexidine-alcohol (0.5%) was used for scrubbing and disinfecting the venipuncture sites and blood culture bottle heads. Venous blood (10 mL) was withdrawn from the peripheral vein using sterile syringes. One set of blood culture tests consisted of two bottles, an aerobic resin bottle (FA Plus) and an anaerobic resin bottle (FN Plus), of the BacT/Alert 3D System. An aliquot of approximately 5 mL of blood was directly inoculated into each bottle. The blood culture bottle weight was measured upon receipt, after blood collection. The obtained weight was divided by the blood density (1.055 g/mL) to determine the blood volume.
Laboratory Procedures
The received bottles were quickly inserted into the automated blood culture system. Bottles that arrived outside of routine business hours were stored at room temperature and were processed the following morning. The bottles were incubated for 5 days unless they were flagged as positive. Bottles with a positive signal were removed from the automated blood culture system. Aliquots (100 µL) were withdrawn to perform gram staining and to inoculate blood agar plates (BAP), MacConkey agar plates, and chocolate agar plates. The agar plates were incubated for 48 hr at 37℃ with 5% CO
2. Anaerobic culturing was selectively conducted when no growth was observed in aerobic culture but when the presence of organisms was confirmed by gram stain. The colonies were identified with the Vitek II System (bioMérieux Inc.). All signal-negative bottles were subjected to terminal subculture after 5 days of incubation. Broth and blood mixture (100 µL) was inoculated onto BAP and incubated for 2 days at 37℃ with 5% CO
2. When more than two microorganisms were recovered from one bottle, it was considered a polymicrobial infection. The positive rate was defined as the number of sets with growth of clinically significant pathogens in either FA Plus or FN Plus, among the requested sets. The positive rate was compared between FA Plus and FN Plus. The contamination rate was defined as the growth of skin contaminants among the requested sets. Growth of
Bacillus spp.,
Corynebacterium spp.,
Propionibacterium spp., and
Micrococcus spp. was regarded as skin contamination, regardless of a positive result [
19]. TTD was defined as the time interval between the entry of the bottles and a positive signal in the BacT/Alert 3D system.
Assessment of Clinical Significance
Upon microorganism identification, the corresponding case was reviewed for medical history, laboratory findings, and suspected diagnosis using an electronic medical record system. The identified microorganisms were categorized as either a pathogen or a contaminant, based on medical history, vital signs, disease progression, treatment response, and laboratory tests such as leukocyte counts, C-reactive protein, lactate dehydrogenase, blood gas analysis, and microbiological culture results from other specimens [
2].
Effect of Prior Antibiotic Use
Medical records were reviewed to determine antibiotic use within 3 days of blood culture tests. If needed, an experienced nurse from the ED called the referred clinic for the history of antibiotic use. The pathogen detection rate and the TTD were compared between prior antibiotic users and antibiotic naïve patients.
Statistical Analysis
The statistical significance regarding positive rates was examined by χ2-test using exact binomial probability calculations as described by McNemar. The normality of the distribution of TTD was assumed by the Kolmogorov-Smirnov test. When TTD data presented the assumption of normality, we used a paired t-test to compare differences in mean TTD between two conditions. Otherwise, the Wilcoxon signed-rank test was applied. All statistical analyses were performed using SPSS, version 21 (IBM Corp., Armonk, NY, USA), and statistical significance was defined as a P value less than 0.05.
RESULTS
Blood Volume
During the study period, 2,994 blood culture sets were submitted to the laboratory. The mean (±SD) blood volume of the received bottles was 4.76 (±1.39) mL in FA Plus and 4.90 (±1.26) mL in FN Plus.
Positivity
Among 2,994 sets received, 371 sets (12.4%) yielded 385 clinically significant pathogens. Using FA Plus and FN Plus, 306 (10.2%) and 305 (10.1%) bottles, respectively, were positive (
P >0.05). Polymicrobial infections were observed in 34 cases (0.1%). When comparing FA Plus to FN Plus, gram-negative lactose non-fermenters (18 vs. 1) and yeasts (15 vs. 0) were isolated at a significantly higher proportion in FA Plus (
P<0.001), whereas gram-positive bacteria (10 vs. 26;
P =0.011) and anaerobes (0 vs. 10;
P<0.01) were more commonly isolated in FN Plus. Gram-negative bacilli were isolated equally (41 in FA Plus vs. 40 in FN Plus) (
Table 1).
Time to Detection
The mean TTD of all pathogens was slightly shorter in FA plus than in FN Plus (11.12 hr vs. 11.88 hr;
P =0.009) (
Table 2). FA Plus resulted in earlier detection of gram-positive cocci and
Staphylococcus aureus compared to its anaerobic counterpart (
P<0.001). FN Plus resulted in an earlier mean TTD in the detection of enterococci, gram-negative bacilli,
Escherichia coli,
Klebsiella pneumoniae, other family members of
Enterobacteriaceae, and yeasts, compared to FA Plus; however, the differences were not statistically significant. The TTD of gram-negative bacilli was the shortest followed by gram-positive cocci and yeasts. The differences in TTD values were significant among these groups (
P<0.05). The distribution of the TTD values was noteworthy (
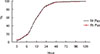
Fig. 1). Over 88% of positive signals were observed in the first 24 hr. More than 97% were detected in both FA and FN bottles within 48 hr. Within 72 h, 99.7% were detected in FA Plus and 99.0% in FN Plus.
Terminal Subculture
After terminal subculture, growth was observed in only nine sets (0.34%) among the 2,623 sets that showed a negative signal for 5 days. They consisted of one (0.04%) FA Plus and eight (0.3%) FN Plus cultures (data not shown). An isolate of Pseudomonas aeruginosa demonstrated growth from the FN Plus bottle, whereas the corresponding FA Plus bottle showed no growth. The other isolates grown by terminal subculture were identical to those of the companion bottle from the same sets.
Contamination Rate
Isolates recovered from 63 sets (2.11%) were assessed as skin contaminants. Skin contaminants were isolated 48 (1.61%) times in FA Plus and 36 (1.21%) times in FN Plus. Coagulase-negative staphylococci (CoNS) were most common (71.4%), followed by Bacillus spp. (20.2%), and Micrococcus spp. (4.8%) (data not shown). Based on chart review and the results of other laboratory tests, we determined eight cases of CoNS to be pathogenic. Given that all CoNS were included as contaminants, the contamination rate rose to 1.71 % in FA Plus, 1.44% in FN Plus, and 2.38% in total.
Effect of Prior Antibiotic Therapy
In total, 249 patients (498 of 2,994 sets, 33.2%) received antibiotics before blood collection. Prior antibiotic therapy was observed in 43.9% (160/364 sets) of MICU cases, 48.2% (54/112 sets) of SICU cases, and 11.2% (284/2,518 sets) of ED cases. The positive rates in prior antibiotic users were 16.3% (26/160) in the MICU, 14.8% (8/54) in the SICU, and 10.9% (65/498) in the ED. The positive rates of antibiotic naïve patients were 16.2% (33/204) in the MICU, 17.2% (10/58) in the SICU, and 11.8% (263/2,234) in the ED. There was no statistically significant difference in the positivity between prior antibiotic users (13.1%, 65/498) and antibiotic naïve patients (12.3%, 306/2,496) (P >0.05). In antibiotic users, TTD was 11.70±6.74 hr in FA Plus and 11.98±5.71 hr in FN Plus. In the antibiotic naïve patients, TTD was 10.96±7.56 hr in FA Plus and 11.84±9.85 hr in FN Plus. The differences in TTD values between these two groups were not statistically significant (P =0.420 for FA Plus, P =0.166 for FN Plus).
DISCUSSION
The recently introduced BacT/Alert 3D resin media were reported to be superior to conventional media. The resin media showed improved detection of gram-negative bacilli compared to standard non-resin containing media [
10].
S. aureus and CoNS were isolated at a higher frequency in the resin media compared to the charcoal media [
11]. In addition, BACTEC Plus resin media showed superiority to BacT/Alert charcoal media for the isolation of organisms [
612]. There have been many clinical studies comparing other blood culture systems (bioMerieux BacT/Alert 3D, Becton Dickinson FX or BACTEC 9600, and Thermo Fisher Versa-TREK) or different media (standard media, charcoal media, resin media, and lysis media) [
61011]. As expected, lactose non-fermenters and fungi grew predominantly in FA Plus because these organisms are absolute aerobes. In contrast, gram-positive organisms and anaerobes grew significantly more in FN Plus. In this study, there was no difference in the isolation of gram-negative organisms between the two media.
Blood volume is a key parameter of blood cultures. Although 20-30 mL of venous blood is recommended for each venipuncture [
13], the currently available commercial blood culture manufacturers recommend inoculating up to 10 mL for each bottle. That recommendation might be due to reduced broth volume and a desirable blood:broth ratio (5-10:1). Therefore, the required blood collection guidelines need to be reviewed by experts. In contrast, many studies have determined that the average blood volume inoculated per bottle is less than 5 mL [
4513]. Thus, we adopted a 5-mL protocol for each bottle. The positive rate in our study was 12.4%, excluding skin contaminants, which was deemed to be satisfactory considering that a desirable positive rate is 6% to 12% [
3]. However, severity of illness in the patient groups needs to be considered to interpret this result. Our study groups (from the ED and ICUs) might include patients with relatively severe conditions compared to those of other patient groups. Positivity was not affected by prior antibiotic use. This result could be explained by the function of the resin, which may adsorb antibiotics effectively.
Although there have been several reports advocating selective use of anaerobic bottles by clinical indications [
14], the results of this protocol are inconclusive. Although bacteremia due to absolute anaerobes has decreased recently [
115], our data showed an equivalent recovery of organisms in anaerobic bottles. Instead of an aerobic/aerobic or aerobic/selective anaerobic combination, we recommend an aerobic/anaerobic set for blood cultures as a standard protocol [
141617].
Gram-positive cocci including
S. aureus grew significantly faster in FA Plus than in FN Plus. Although there were statistically significant differences in TTD values for these microorganisms between FA Plus and FN Plus, the mean TTD values for all micro-organisms were very similar (11.1 hr for FA Plus, 11.9 hr for FN Plus). Since more than 97.0% of positives were detected within 48 hr and more than 99.0% within 72 hr, laboratories with limited resources could apply a 3-day incubation protocol instead of a 5-day incubation. This was consistent with a previous study using a different instrument [
18].
Terminal subculture was performed for all signal-negative bottles. Microorganism growth by terminal subculture was more common from anaerobic bottles. From terminal subculture, nine cases were positive. Among them, three samples were processed during the night shift. The specimens were stored at room temperature until the next morning (for 5, 9, 15 hr, respectively). This delayed entry might have affected growth after subculture. In eight cases, microorganisms had already been detected from companion bottles. Only one isolate of P. aeruginosa grew from a set showing a negative signal. This patient was diagnosed as having a urinary tract infection, and P. aeruginosa was isolated from the urine culture. She survived after optimal management. Taken together, we conclude that terminal subculture would not be necessary for resin media of BacT/Alert 3D.
A desirable skin contamination rate is <3% [
3]. Sometimes classification of the isolate as a true pathogen or a contaminant is difficult, especially with CoNS [
1915]. An experienced ED physician reviewed the medical records and categorized the isolates into two groups, clinically significant or contaminant. The skin contamination rate was 2.1%, and there was no significant difference in the contamination rate between FA Plus and FN Plus.
Serious limitations of our study are the small inoculum of blood and delay of entry into the machine during the night shift hours. The differentiation of pathogens and skin contaminants was rather subjective. The order of inoculation occurred most often as, FA Plus first, followed by FN Plus. This order may affect the positive rate and contamination rate of each bottle. However, many experts recommend inoculating the aerobic bottle first [
39]. Although blood culture bottles should enter the blood culture machine within 2 hr of collection [
3], occasional delayed entry is inevitable. The clinical microbiology laboratory is closed during the night at our hospital and is a significant distance from the stat laboratory. Therefore, we placed the bottles that arrived during the night at room temperature until the following morning. Many studies on delayed entry argued that a harmful effect would be minimal for up to 24 hr of storage at room temperature [
91719]. We found that only one-third (3/9) of growth by terminal subculture was from sets that arrived during the night shift. This suggests that delayed entry might not be the major reason for false negatives in the resin media of the BacT/Alert 3D system.
In conclusion, clinical performance of the BacT/Alert 3D resin media showed an excellent positive rate and low contamination rate. Detection of microorganisms was complementary between FA Plus and FN Plus. Positive rates were the same between these two media, and were not affected by prior antibiotic treatment. In addition, terminal subculture does not seem necessary, because the false-negative rate was negligible. Gram-positive cocci including S. aureus grew significantly earlier in FA Plus than in FN Plus.






 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download