Abstract
Background
We aimed to conduct a systematic review of previously published material to evaluate the diagnostic effectiveness of PCR-based tests in detecting BRAF mutation.
Methods
Eight Korean databases, including KoreaMed, Ovid-MEDLINE, and Ovid-EMBASE were used to identify relevant published studies. Nine studies describing usage of real-time PCR, dual-priming oligonucleotide (DPO)-multiplex real-time PCR and allele-specific PCR were included in the final assessment. Two reviewers screened all references independently for assessing the quality of the included articles and extracted data.
Results
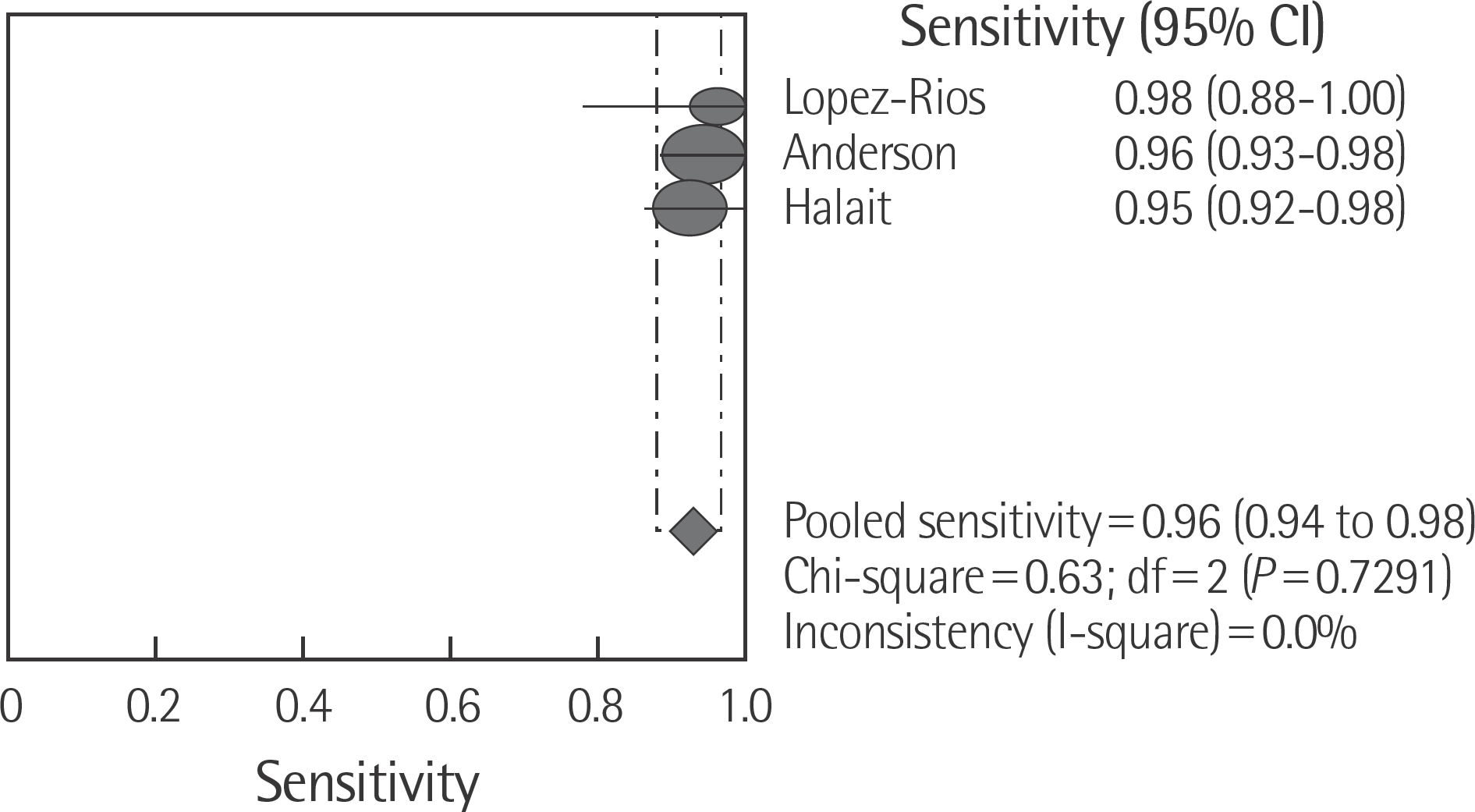
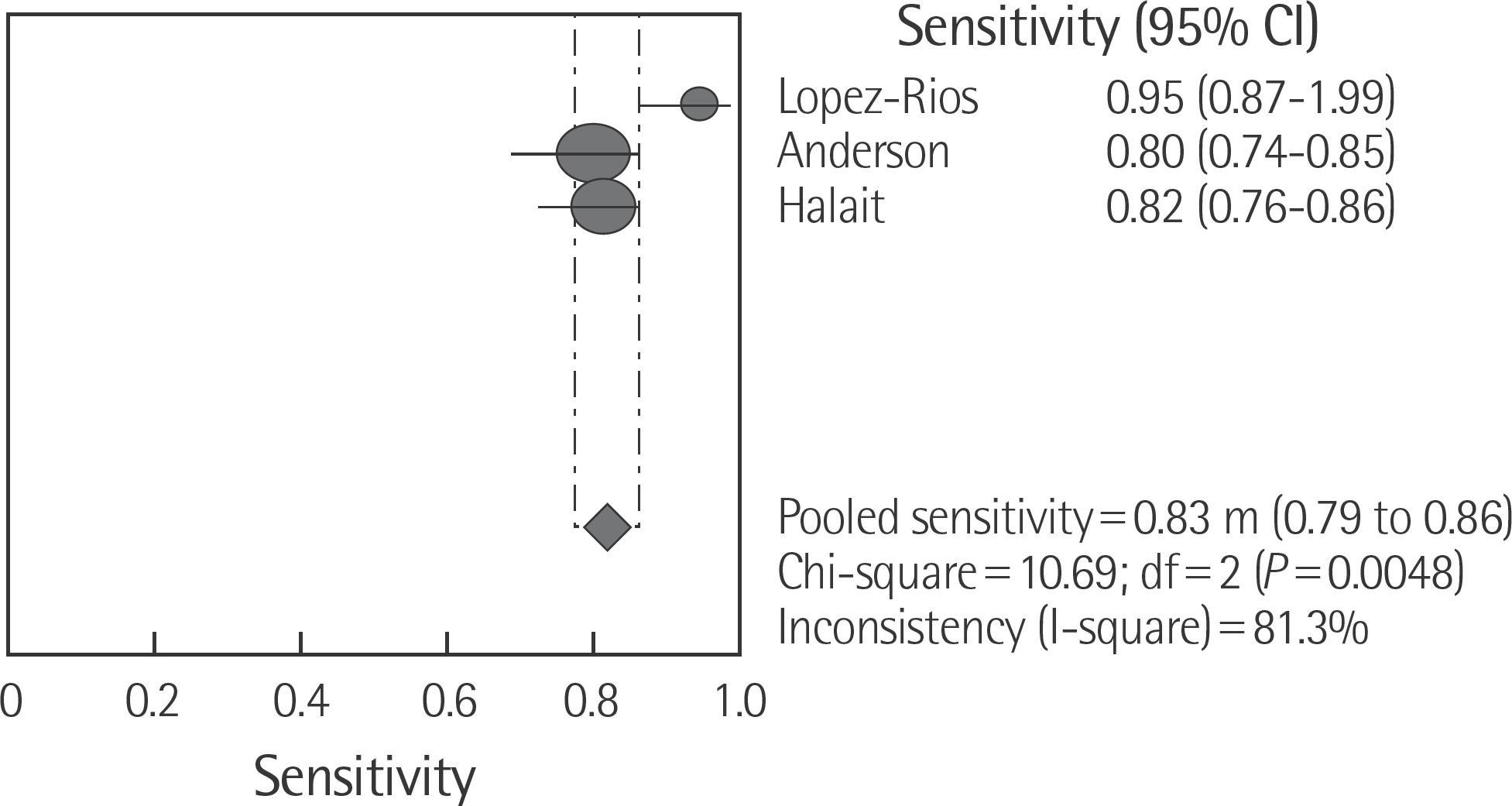
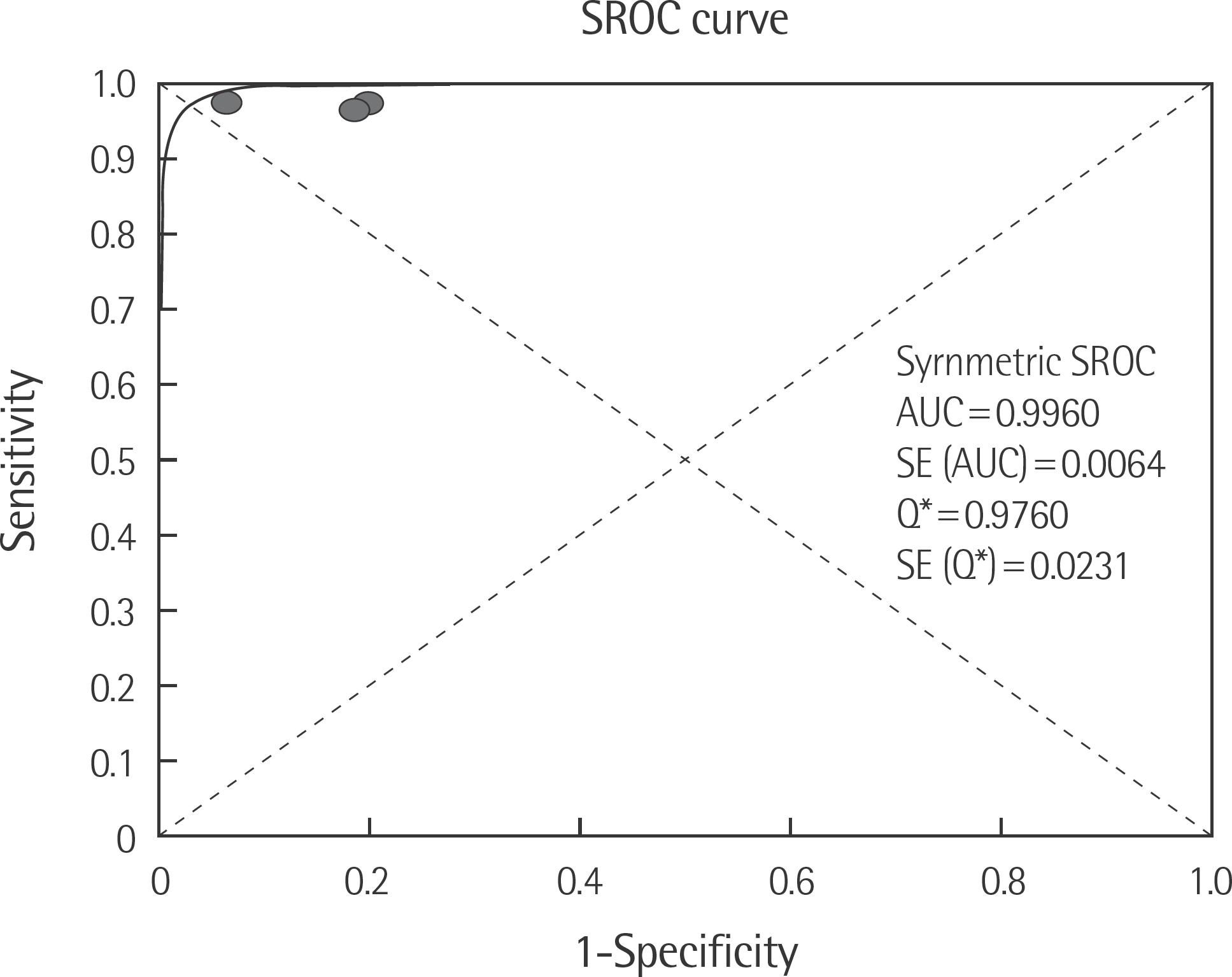
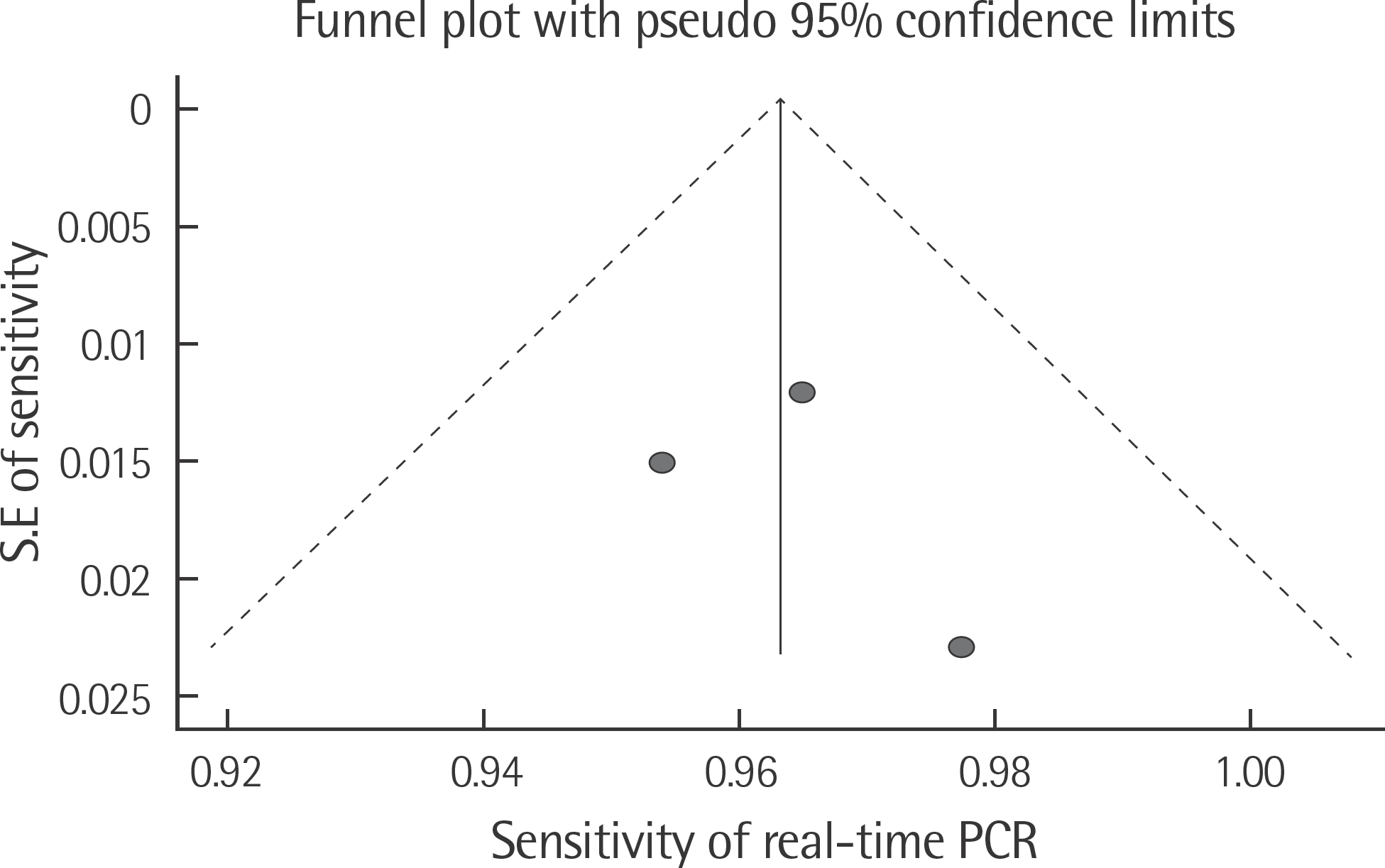
The rate of detection of the BRAF mutations was lower in the Korean population (11.1–17.2%) than that in the Western population (36.7–82.2%). The diagnostic accuracy of the BRAF mutation tests was assessed on the basis of four previous reports, all of which employed real-time PCR on malignant melanoma. In fact, the diagnostic accuracy of real-time PCR was found to be higher than that of sequencing tests (pooled sensitivity, 0.96; pooled specificity, 0.83; and summary receiver operating characteristic area under the curve, 0.99). In addition, we found that there was no publication bias in meta-analysis. The concordance rate of the BRAF mutation tests compared with reference tests was 87.9–98.1%.
Go to : 
References
1. Rha SY, Lee JC, Kwon KH, Lee HJ, Kim KS, Jo YS, et al. The relationship between the BRAF mutation in thyroid papillary carcinomas and the prognostic factors. J Korean Soc Endocrinol. 2005; 20:224–9.
2. National Comprehensive Cancer Network (NCCN) Guidelines. http://www.nccn.com. (Updated on Dec 2013).
3. National Institute for Health and Clinical Excellence (NICE) technology appraisal guidance 269, Vemurafenib for treating locally advanced or metastatic BRAF V600 mutation-positive malignant melanoma. December 2012.
4. Korean Society for Labortory Medicine. Park CJ, ed. Laboratory Medicine. 4th ed.Reston: E∗PUBLIC;2009. p. 828–9.
5. Venesio T, Chiorino G, Balsamo A, Zaccagna A, Petti C, Scatolini M, et al. In melanocytic lesions the fraction of BRAF V600E alleles is associated with sun exposure but unrelated to ERK phosphorylation. Mod Pathol. 2008; 21:716–26.
6. SIGN 50: a guideline developer's handbook. Scottish Intercollegiate Guidelines Network. http://www.sign.ac.uk/pdf/sign50.pdf. (Updated on Jan 2013).
7. Lopez-Rios F, Angulo B, Gomez B, Mair D, Martinez R, Conde E, et al. Comparison of testing methods for the detection of BRAF V600E mutations in malignant melanoma: preapproval validation study of the companion diagnostic test for vemurafenib. PLoS ONE. 2013; 8:e53733.

8. Anderson S, Bloom KJ, Vallera DU, Rueschoff J, Meldrum C, Schilling R, et al. Multisite analytic performance studies of a Real-Time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed paraffin embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med. 2012; 136:1385–91.
9. Halait H, De Martin K, Shah S, Soviero S, Langland R, Cheng S, et al. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF gene, used as the companion diagnostic test for the novel BRAF inhibitor Vemurafenib in metastatic melanoma. Diagn Mol Pathol. 2012; 21:1–8.

10. Schoenewolf NL, Dummer R, Mihic-Probst D, Moch H, Simcock M, Ochsenbein A, et al. Detecting BRAF mutations in formalin-fixed melanoma: experiences with twostate-of-the-art techniques. Case Rep Oncol. 2012; 5:280–9.
11. Lee HW, Song KH, Hong JW, Jeon SY, Ko DY, Kim KH, et al. Frequc-ney of BRAF mutation and clinical relevance for primary melanomas. Korean J Pathol. 2012; 46:246–52.
12. Hacker E, Hayward NK, Dumenil T, James MR, Whiteman DC. The association between MC1R genotype and BRAF mutation status in cutaneous melanomas: findings from an Australian population. J Invest Dermatol. 2010; 130:241–8.
13. Lazar V, Ecsedi S, Attila G Szöllo˝si, Tóth R, Vízkeleti L, Rákosy Z, et al. Characterization of candidate gene copy number alterations in the 11q13 region along with BRAF and NRAS mutations in human melanoma. Mod Pathol. 2009; 22:1367–78.

14. Liu W, Kelly JW, Trivett M, Murray WK, Dowling JP, Wolfe R, et al. Distinct clinical and pathological features are associated with the BRAFT1799A(V600E) mutation in primary melanoma. J Invest Dermatol. 2007; 127:900–5.
15. Kim JO, Yang HY, Son S, Park KH. Clinical experiences in malignant melanoma. J Korean Surg Soc. 1997; 53:905–10.
16. National Cancer Information Center. http://cancer.go.kr. (Updated on Dec 2013).
17. Sung WJ, Park SJ, Gu MJ, Bae YK. Automated silver-enhanced in situ hybridization for evaluation of HER2 gene status in breast carcinoma: comparison with fluorescence in situ hybridization and immunohistochemistry. Korean J Pathol. 2010; 44:28–34.

18. National Cancer Institute (NCI). http://www.cancer.gov. (Updated on Dec 2013).
19. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. BRIM-3 Study Group. Improved survival with Vemurafenib in melanoma with BRAF V600E mutation. New Engl J Med. 2011; 364:2507–16.
20. Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New Engl J Med. 2010; 363:809–19.

21. Cunnon B and May JW Jr. Skin contractures of the hand. Flynn JE, editor. Hand surgery. 2nd ed.Baltimore: Williams and Wilkins;2006. p. 776–7.
22. McGovern VJ. The nature of melanoma. A critical review. J Cutan Pathol. 1982; 9:61–81.
23. Jin SA, Chun SM, Choi YD, Kweon SS, jung ST, Shim HJ, et al. BRAF mutations and KIT aberrations and their clinicopathological correlation in 202 Korean melanomas. J Invest Dermatol. 2013; 133:579–82.

24. Ziai J, Hui P. BRAF mutation testing in clinical practice. Expert Rev Mol Diagn. 2012; 12:127–38.
25. Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003; 33:19–20.

26. Scottish Intercollegiate Guidelines Network (SIGN). URL:. http://www. sign.ac.uk (Updated on Dec 2013).
Go to : 
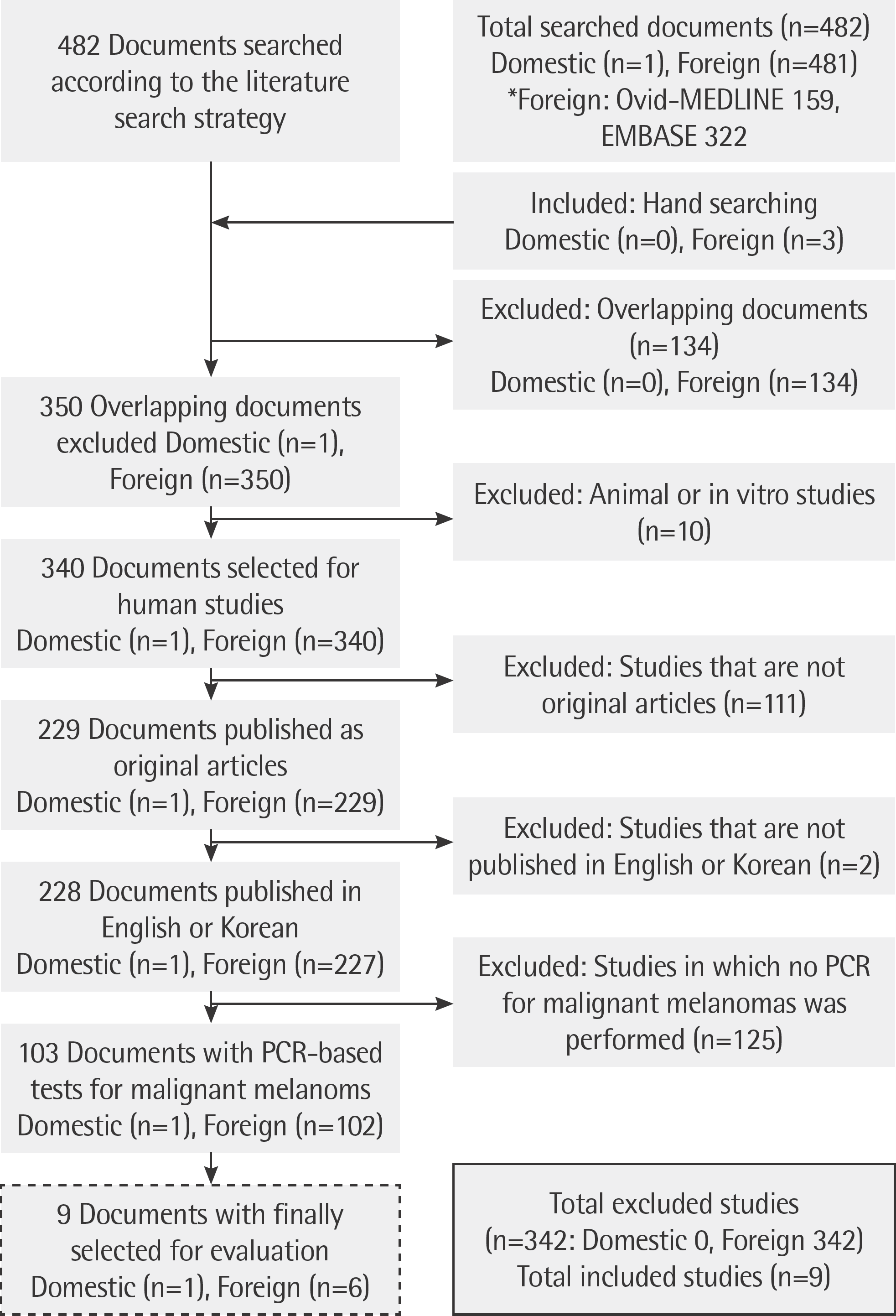 | Fig. 1.Documents selected for evaluation of PCR-based BRAF mutation test according to the literature search strategy. |
Table 1.
Databases used for searching published studies
Table 2.
Ovid-MEDLINE search strategy
Table 3.
Ovid-EMBASE search strategy
Table 4.
Levels of evidence (From Scottish Intercollegiate Guidelines Network. SIGN 50: a guideline developer's handbook)
Table 5.
Grades of recommendations (From Scottish Intercollegiate Guidelines Network. SIGN 50: a guideline developer's handbook)
Table 6.
Documents selected for evaluation and detection rate of the BRAF mutation
| No. | Author/Publication Year | Research location | Subjects | Index test | Detection rate %(N) | BRAF mutation type (N) | Level of evidence |
|---|---|---|---|---|---|---|---|
| 1 | Lopez-Rios et al. (2013) [7] | U.S | 108 melanoma | Real-Time PCR | 39.8 (43/108) | V600E | 2+ |
| FFPET specimens∗ | |||||||
| 2 | Anderson et al. (2012) [8] | US, Australia | 477 metastatic | Real-Time PCR | 49.6 (215/433) | V600E | 2+ |
| melanoma patients | |||||||
| 3 | Halait et al. (2012) [9] | U.S | 438 malignant | Real-Time PCR | 43.1 (189/438) | V600E | 2+ |
| melanoma FFPET | |||||||
| specimens∗ | |||||||
| 4 | Schoenewolf et al. (2012) [10] | Swiss | 62 unresectable stage | Real-Time PCR | 34.6 (18/52)† | V600E | 2− |
| IV melanomas | |||||||
| 5 | Lee et al. (2012) [11] | South Korea | 58 primary | DPO-multiplex | 17.2 (10/58) | – | 2− |
| cutaneous melanomas | Real-Time PCR | ||||||
| 27 non-cutaneous | 11.1 (3/27) | ||||||
| melanomas | |||||||
| 6 | Hacker et al. (2010) [12] | Australia | 123 melanomas | AS-PCR | 32.5 (40/123) | V600 | 2− |
| 7 | Lazar et al. (2009) [13] | Hungary | 68 primary melanoma | Real-Time Quantitative PCR | 39.7 (27/68) | V600E (20/68) V600K (5/68) | 2+ |
| V600R (2/68) | |||||||
| 8 | Venesio et al. (2008) [5] | Italy | 18 cutaneous melanoma | AS-PCR | 82.2 (13/18) | V600E | 2+ |
| 9 | Liu et al. (2007) [14] | Australia | 251 invasive | AS-PCR | 44.6 (112/251) | V600E | 2− |
| primary melanomas | (T1799A) |
Table 7.
Diagnostic concordance rate




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download