Abstract
A new clinico-pathological entity in which isochromosome 17q is the sole abnormality has been reported in myelodysplastic syndrome and in myeloproliferative neoplasm with an aggressive course; In particular, myelodysplastic syndrome with the isolated i(17)(q10) chromosome has the unique features of male sex, severe anemia, dysmegakaryocytic hyperplasia, increased micromegakaryocytes, basophilia, eosinophila and high risk for progression to acute myeloid leukemia (AML). However, the isolated i(17)(q10) is occurring at a relatively low frequency in de novo AML, and only a few reports are available in the literature about the clinical features and molecular characteristics of the isolated i(17)(q10) in AML. Herein, we report both the clinico-pathological features and the results of high resolution single nucleotide polymorphism (SNP) array analysis in a case of AML with i(17)(q10) as the sole cytogenetic abnormality. This case showed marrow findings of basophilia and dysmegakaryocytic hyperplasia and aggressive clinical outcome and these findings were suggestive of the presence of underlying myelodysplastic syndrome. The breakpoint of i(17)(q10) was located within 17p11.2 sub-band, which is known as a genetically highly unstable region presenting a unique genomic architectural features of low copy repeats (LCRs); thus, LCRs within 17p11.2 might lead to genomic instability and facilitate somatic genetic rearrangements such as i(17) (q10) and could play an important pathogenetic role in presenting unique clinico-pathologic features as well as in tumor development and disease progression.
Figures and Tables
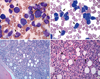 | Fig. 1Bone marrow aspiration and biopsy findings. Bone marrow aspiration showing marked proliferation of leukemic blasts (68%) with occasional unusual cytoplasmic basophilc granules, and increase of basophils (10%) indicated by arrows (Wright-Giemsa stain, ×1,000, A and B). Bone marrow biopsy showing hypercellularity with leukemic blasts and dysplastic megakaryocytes including micromegakaryocytes indicated by arrows (H&E, ×200 and ×400, C and D). |
 | Fig. 2Bone marrow biopsy showing scattered positive cells for c-KIT (CD117) and those for tryptase. (A) Immunohistochemical stain, c-KIT (×200). (B) Immunohistochemical stain, tryptase (×400). |
 | Fig. 3Giemsa-banding karyotype analysis revealed 46,XY,i(17)(q10) in 19 out of 20 metaphases examined. |
References
1. Solé F, Torrabadella M, Granada I, Florensa L, Vallespi T, Ribera JM, et al. Isochromosome 17q as a sole anomaly: a distinct myelodysplastic syndrome entity? Leuk Res. 1993. 17:717–720.

2. Fioretos T, Strömbeck B, Sandberg T, Johansson B, Billström R, Borg A, et al. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood. 1999. 94:225–232.

3. Xiao Z, Liu S, Yu M, Xu Z, Hao Y. Isochromosome 17q in patients with myelodysplastic syndromes: six new cases. Haematologica. 2003. 88:714–715.
4. Becher R, Carbonell F, Bartram CR. Isochromosome 17q in Ph1-negative leukemia: a clinical, cytogenetic, and molecular study. Blood. 1990. 75:1679–1683.

5. Nishida H, Ueno H, Park JW, Yano T. Isochromosome i(17q) as a sole cytogenetic abnormality in a case of leukemic transformation from myelodysplastic syndrome (MDS)/myeloproliferative diseases (MPD). Leuk Res. 2008. 32:1325–1327.

6. Soenen V, Preudhomme C, Roumier C, Daudignon A, Laï JL, Fenaux P. 17p Deletion in acute myeloid leukemia and myelodysplastic syndrome. Analysis of breakpoints and deleted segments by fluorescence in situ. Blood. 1998. 91:1008–1015.

7. Pinheiro RF, Chauffaille Mde L, Silva MR. Isochromosome 17q in MDS: a marker of a distinct entity. Cancer Genet Cytogenet. 2006. 166:189–190.

8. Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002. 21:3475–3495.

9. Lazarević V, Djordjević V, Magić Z, Marisavljevic D, Colović M. Refractory anemia with ring sideroblasts associated with i(17q) and mutation of the TP53 gene. Cancer Genet Cytogenet. 2002. 136:86–89.

10. Peetre C, Nilsson PG, Mitelman F. Isochromosome 17q in a patient with acute myeloblastic leukemia. Cancer Genet Cytogenet. 1987. 24:315–318.

11. Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999. 14:1–8.

12. Kanagal-Shamanna R, Bueso-Ramos CE, Barkoh B, Lu G, Wang S, Garcia-Manero G, et al. Myeloid neoplasms with isolated isochromosome 17q represent a clinicopathologic entity associated with myelodysplastic/myeloproliferative features, a high risk of leukemic transformation, and wild-type TP53. Cancer. 2012. 118:2879–2888.

13. Yi JH, Huh J, Kim HJ, Kim SH, Kim HJ, Kim YK, et al. Adverse prognostic impact of abnormal lesions detected by genome-wide single nucleotide polymorphism array-based karyotyping analysis in acute myeloid leukemia with normal karyotype. J Clin Oncol. 2011. 29:4702–4708.

14. Makishima H, Rataul M, Gondek LP, Huh J, Cook JR, Theil KS, et al. FISH and SNP-A karyotyping in myelodysplastic syndromes: improving cytogenetic detection of del(5q), monosomy 7, del(7q), trisomy 8 and del(20q). Leuk Res. 2010. 34:447–453.

15. Tiu RV, Gondek LP, O'Keefe CL, Elson P, Huh J, Mohamedali A, et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood. 2011. 117:4552–4560.

16. Bullinger L, Krönke J, Schön C, Radtke I, Urlbauer K, Botzenhardt U, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010. 24:438–449.

17. Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009. 27:5219–5226.

18. Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005. 65:9152–9154.

19. Koren-Michowitz M, Sato-Otsubo A, Nagler A, Haferlach T, Ogawa S, Koeffler HP. Older patients with normal karyotype acute myeloid leukemia have a higher rate of genomic changes compared to young patients as determined by SNP array analysis. Leuk Res. 2012. 36:467–473.

20. Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Höglund M, et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004. 74:1–10.

21. Carvalho C, Lupski JR. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008. 18:1724–1732.

22. Shin I, Kim S, Song H, Kim HR, Moon A. H-Ras-specific activation of Rac-MKK3/6-p38 pathway: its critical role in invasion and migration of breast epithelial cells. J Biol Chem. 2005. 280:14675–14683.
24. Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009. 106:12950–12955.

25. Suela J, Alvarez S, Cifuentes F, Largo C, Ferreira BI, Blesa D, et al. DNA profiling analysis of 100 consecutive de novo acute myeloid leukemia cases reveals patterns of genomic instability that affect all cytogenetic risk groups. Leukemia. 2007. 21:1224–1231.

26. Fiedler W, Weh HJ, Hegewisch-Becker S, Hossfeld DK. GCSF gene is expressed but not rearranged in a patient with isochromosome 17q positive acute nonlymphocytic leukemia. Cancer Genet Cytogenet. 1993. 68:49–51.

27. Pozdnyakova O, Miron PM, Tang G, Walter O, Raza A, Woda B, et al. Cytogenetic abnormalities in a series of 1,029 patients with primary myelodysplastic syndromes: a report from the US with a focus on some undefined single chromosomal abnormalities. Cancer. 2008. 113:3331–3340.

28. Nowell P, Finan J. Chromosome studies in preleukemic states. IV. Myeloproliferative versus cytopenic disorders. Cancer. 1978. 42:2254–2261.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download