Abstract
Herein, we report a case in which cholestasis caused discrepancy in high-density lipoprotein (HDL)-cholesterol levels measured using various methods. The discrepancy in HDL-cholesterol level originated from the abnormal increase in the level of an unusual lipoprotein, apo E-rich HDL, in the patient's serum. An abnormal slow alpha-migrating lipoprotein was observed on agarose gel electrophoresis, and an abnormal large-sized HDL was observed in a lipoprotein subfraction study. The level of apolipoprotein E was elevated.
Figures and Tables
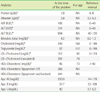
Fig. 2
LDL subfraction test result of the patient.
The red circle shows the large-sized abnormal HDL fraction (b).

References
1. Min WK, Park H, Jun KR, Chun S, Lee W. Factors affecting the difference between the low-density lipoprotein cholesterol concentrations measured directly and calculated using the friedewald formula. J Lab Med Qual Assur. 2008. 30:233–235.
2. Hur M, Kim CS, Park MJ, Kwak I, Lee KM. Analysis of low-density lipoprotein cholesterol by homogenous assay in comparison with friedewald formula. Korean J Lab Med. 2003. 23:104–108.
3. Remaley AT, Rifai N, Warnick R. Burtis CA, Ashwood ER, editors. Lipids, lipoproteins, apolipoproteins, and other cardiovascular risk factors. Tietz textbook of clinical chemistry and molecular diagnostics. 2012. 5th ed. St. Louis: Elsevier;731–805.

4. Remaley AT, Warnick GR. HDL-C. The changing testing paradigm part 1. Clinical Laboratory News. 2007. Washington: American Association for Clinical Chemistry.
5. Clifton PM, Barter PJ, Mackinnon AM. High density lipoprotein particle size distribution in subjects with obstructive jaundice. J Lipid Res. 1988. 29:121–135.

6. Coulhon MP, Tallet F, Yonger J, Agneray J, Raichvarg D. Changes in human high density lipoproteins in patients with extra-hepatic biliary obstruction. Clin Chim Acta. 1985. 145:163–172.

7. Yamashita S, Sprecher DL, Sakai N, Matsuzawa Y, Tarui S, Hui DY. Accumulation of apolipoprotein E-rich high density lipoproteins in hyperalphalipoproteinemic human subjects with plasma cholesteryl ester transfer protein deficiency. J Clin Invest. 1990. 86:688–695.

8. Chiba H, Osaka T, Iwasaki N, Suzuki H, Akizawa K, Fujisawa S, et al. Spurious elevation of serum high-density lipoprotein cholesterol in patients with cholestatic liver diseases. Biochem Med Metab Biol. 1991. 46:329–343.

9. Tallet F, Vasson MP, Couderc R, Lefevre G, Raichvarg D. Characterization of lipoproteins during human cholestasis. Clin Chim Acta. 1996. 244:1–15.

10. Black DD. Chronic cholestasis and dyslipidemia: what is the cardiovascular risk? J Pediatr. 2005. 146:306–307.

11. Werner A, Kuipers F, Verkade HJ. Fat absorption and lipid metabolism in cholestasis. Madame Curie Bioscience Database [Internet]. 2000. Austin (TX): Landes Bioscience.

12. Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002. 109:961–971.

13. Lund-Katz S, Phillips MC. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell Biochem. 2010. 51:183–227.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download