Abstract
Background
Accumulation of genetic aberrations in MDS is closely associated with progression to AML. FLT3-ITD is commonly found in AML and less frequently in MDS. FLT3-ITD in MDS is associated with a high risk of transformation to AML. Recently, significant interaction of NPM1 and FLT3-ITD was described in AML. This study was conducted to investigate the incidence and prognostic role of FLT3-ITD and NPM1 mutations (NPM1mt) on paired samples at diagnosis of MDS and AML.
Methods
Patients who were diagnosed as MDS transforming to AML were included. FLT3-ITD was detected by PCR, and NPM1mt was confirmed by direct sequencing after screening for NPM by immunohistochemistry.
Results
AML developed in 12.0% (43/357) of MDS patients. FLT3-ITD was detected in none of MDS and 14.7% (5/34) of AML. NPM1mt was detected in 2.4% (1/41) of MDS and 11.6% (5/43) of AML. One patient with type B NPM1mt at MDS transformed to type A NPM1mt at AML. FLT3-ITD positive AML showed a tendency of shorter survival and a significantly longer time to achieve complete remission than FLT3-ITD negative AML (P=0.007). Normal karyotype AML with FLT3-ITD showed shorter overall survival than that group of AML without FLT3-ITD (P=0.017).
Conclusions
MDS patients acquired FLT3-ITD during AML transformation, and FLT3-ITD positive AML, especially that with normal karyotype, predicted a poor outcome. NPM1mt was identified in both MDS and AML. NPM1mt was rarely found in MDS patients, and mostly was acquired after AML transformation. Clonal evolution of NPM1mt subtype was found in one patient during acute transformation.
Go to : 
References
1. Hofmann WK, Lubbert M, Hoelzer D, Phillip Koeffler H. Myelodysplastic syndromes. Hematol J. 2004; 5:1–8.

2. Morgan MA and Reuter CW. Molecularly targeted therapies in myelodysplastic syndromes and acute myeloid leukemias. Ann Hematol. 2006; 85:139–63.

3. Rosnet O, Schiff C, Pebusque MJ, Marchetto S, Tonnelle C, Toiron Y, et al. Human FLT3/FLK2 gene: cDNA cloning and expression in hematopoietic cells. Blood. 1993; 82:1110–9.

4. Shih LY, Lin TL, Wang PN, Wu JH, Dunn P, Kuo MC, et al. Internal tandem duplication of fms-like tyrosine kinase 3 is associated with poor outcome in patients with myelodysplastic syndrome. Cancer. 2004; 101:989–98.
5. Shih LY, Huang CF, Wang PN, Wu JH, Lin TL, Dunn P, et al. Acquisition of FLT3 or N-ras mutations is frequently associated with progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia. 2004; 18:466–75.

6. Yun JP, Chew EC, Liew CT, Chan JY, Jin ML, Ding MX, et al. Nucleo-phosmin/B23 is a proliferate shuttle protein associated with nuclear matrix. J Cell Biochem. 2003; 90:1140–8.

7. Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989; 56:379–90.

8. Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, et al. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989; 28:1033–9.

9. Feuerstein N, Chan PK, Mond JJ. Identification of numatrin, the nuclear matrix protein associated with induction of mitogenesis, as the nucleolar protein B23. Implication for the role of the nucleolus in early transduction of mitogenic signals. J Biol Chem. 1988; 263:10608–12.

10. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005; 352:254–66.

11. Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006; 108:1999–2005.

12. Cazzaniga G, Dell´Oro MG, Mecucci C, Giarin E, Masetti R, Rossi V, et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood. 2005; 106:1419–22.

13. Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002; 100:59–66.

14. Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, Suzuki R, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005; 106:2854–61.

15. Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005; 106:3740–6.
16. Falini B. Any role for the nucleophosmin (NPM1) gene in myelodys-plastic syndromes and acute myeloid leukemia with chromosome 5 abnormalities? Leuk Lymphoma. 2007; 48:2093–5.

17. Ishikawa Y, Xu J, Sakashita G, Urano T, Suzuki T, Tomita A, et al. Abnormal cytoplasmic dyslocalisation and/or reduction of nucleophosmin protein level rarely occurs in myelodysplastic syndromes. Leuk Lymphoma. 2008; 49:2359–64.

18. Shiseki M, Kitagawa Y, Wang YH, Yoshinaga K, Kondo T, Kuroiwa H, et al. Lack of nucleophosmin mutation in patients with myelodysplastic syndrome and acute myeloid leukemia with chromosome 5 abnormalities. Leuk Lymphoma. 2007; 48:2141–4.

19. Zhang Y, Zhang M, Yang L, Xiao Z. NPM1 mutations in myelodysplastic syndromes and acute myeloid leukemia with normal karyotype. Leuk Res. 2007; 31:109–11.

20. Swerdlow SH, Campo E, et al. eds. WHO Classification of tumours of Haematopoietic and Lymphoid Tissues. 4th ed.Lyon: IARC;2008.
21. Vince A, Poljak M, Seme K. DNA extraction from archival Giemsa-stained bone-marrow slides: comparison of six rapid methods. Br J Haematol. 1998; 101:349–51.

22. Kiyoi H, Naoe T, Yokota S, Nakao M, Minami S, Kuriyama K, et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho). Leukemia. 1997; 11:1447–52.
23. Huang Q, Chen W, Gaal KK, Slovak ML, Stein A, Weiss LM. A rapid, one step assay for simultaneous detection of FLT3/ITD and NPM1 mutations in AML with normal cytogenetics. Br J Haematol. 2008; 142:489–92.
24. Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997; 89:2079–88.

25. Bacher U, Schnittger S, Haferlach T. Molecular genetics in acute myeloid leukemia. Curr Opin Oncol. 2010; 22:646–55.

26. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010; 363:2424–33.

27. Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009; 361:1058–66.
28. Steensma DP and Bennett JM. The myelodysplastic syndromes: diagnosis and treatment. Mayo Clin Proc. 2006; 81:104–30.
29. Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009; 114:3448–58.

30. Horiike S, Misawa S, Nakai H, Kaneko H, Yokota S, Taniwaki M, et al. N-ras mutation and karyotypic evolution are closely associated with leukemic transformation in myelodysplastic syndrome. Leukemia. 1994; 8:1331–6.
31. Tien HF, Tang JH, Tsay W, Liu MC, Lee FY, Wang CH, et al. Methylation of the p15 (INK4B) gene in myelodysplastic syndrome: it can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol. 2001; 112:148–54.
32. Horiike S, Yokota S, Nakao M, Iwai T, Sasai Y, Kaneko H, et al. Tandem duplications of the FLT3 receptor gene are associated with leukemic transformation of myelodysplasia. Leukemia. 1997; 11:1442–6.

33. Bacher U, Haferlach T, Kern W, Haferlach C, Schnittger S. A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematologica. 2007; 92:744–52.

34. Georgiou G, Karali V, Zouvelou C, Kyriakou E, Dimou M, Chrisochoou S, et al. Serial determination of FLT3 mutations in myelodysplastic syndrome patients at diagnosis, follow up or acute myeloid leukaemia transformation: incidence and their prognostic significance. Br J Haematol. 2006; 134:302–6.

35. Pinheiro RF, de Sa Moreira E, Silva MR, Alberto FL, Chauffaille Mde L. FLT3 internal tandem duplication during myelodysplastic syndrome follow-up: a marker of transformation to acute myeloid leukemia. Cancer Genet Cytogenet. 2008; 183:89–93.

36. Visani G, Bernasconi P, Boni M, Castoldi G, Ciolli S, Clavio M, et al. The prognostic value of cytogenetics is reinforced by the kind of in-duction/consolidation therapy in influencing the outcome of acute myeloid leukemia–analysis of 848 patients. Leukemia. 2001; 15:903–9.
37. Palmisano M, Grafone T, Ottaviani E, Testoni N, Baccarani M, Martinelli G. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica. 2007; 92:1268–9.

38. Papadaki C, Dufour A, Seibl M, Schneider S, Bohlander SK, Zellmeier E, et al. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: clonal evolution is a limiting factor. Br J Haematol. 2009; 144:517–23.
Go to : 
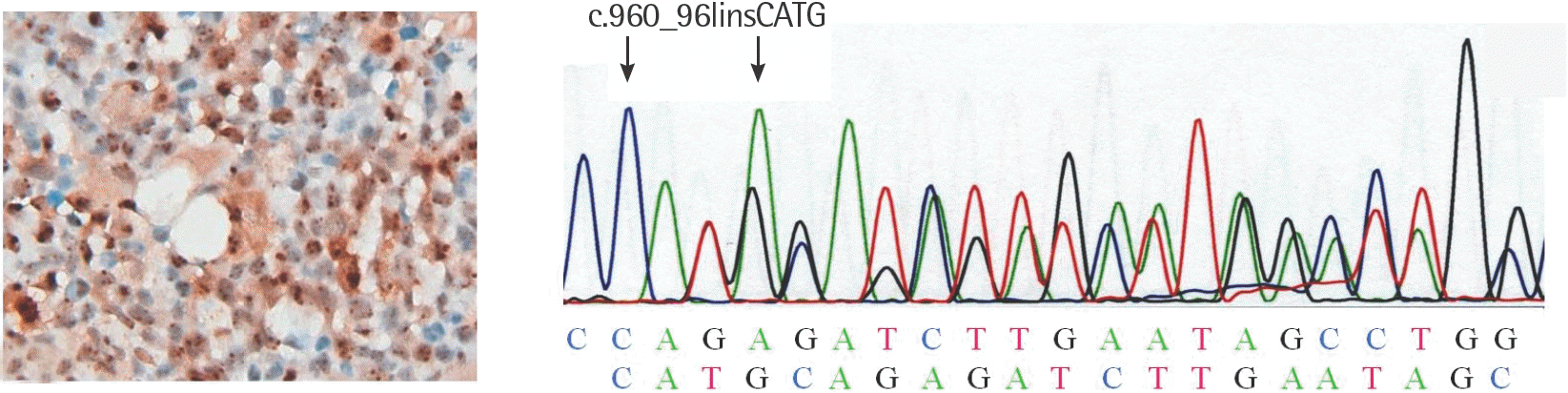 | Fig. 1.Results of NPMc+ and NPM1 exon 12 type B mutation in a patient with AML M2 transformed from RAEB1. (A) Some dysplastic cells show cytoplasmic expression with blurring margination for NPM. (B) Type B mutation of NPM1 with CATG insertion (GenBank Accession No. AY740635) was confirmed by sequencing. Abbreviations: See Tables 1 and 3. |
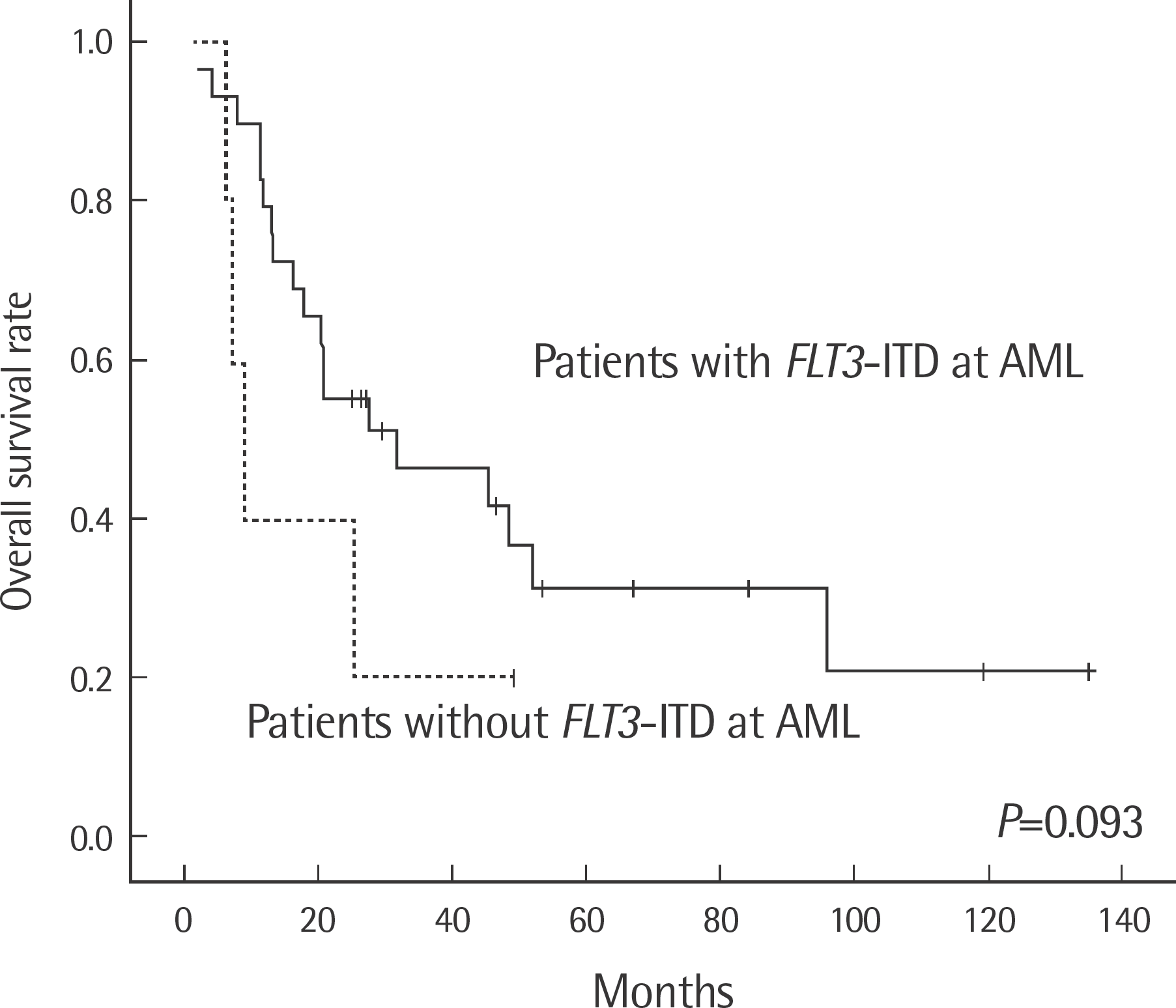 | Fig. 2.Kaplan-Meier survival curves. MDS patients with FLT3-ITD (N=5, straight line) or without FLT3-ITD (N=29, dotted line) at AML transformation. Abbreviations: See Table 1. |
Table 1.
Characteristics of different subtypes of MDS patients according to WHO classification
Table 2.
Results of chromosomal analysis in patients with paired samples at MDS and AML stages
| Age | Sex | Initial diagnosis | Karyotype | |
|---|---|---|---|---|
| MDS | AML | |||
| 41 | M | RA | Normal | Normal |
| 66 | M | RCMD | Normal | Normal |
| 25 | M | RAEB-1 | Normal | Normal |
| 48 | M | RAEB-1 | Normal | Normal |
| 65 | F | RAEB-1 | Normal | Normal |
| 3 | F | RAEB-2 | Normal | Normal |
| 25 | F | RAEB-2 | Normal | Normal |
| 29 | M | RAEB-2 | Normal | Normal |
| 42 | F | RAEB-2 | Normal | Normal |
| 54 | M | RAEB-2 | Normal | Normal |
| 60 | F | RAEB-2 | Normal | Normal |
| 35 | M | RAEB-1 | +8 | Normal∗ |
| 51 | F | RAEB-2 | –7, +21 | –7,+21 |
| 27 | M | RAEB-1 | del (9q) | del (9q) |
| 15 | M | RAEB-2 | t (6;9) | t (6;9) |
| 27 | M | RAEB-2 | t (6;11) | t (6;11) |
| 44 | F | RAEB-2 | t (6;9) | t (6;9) |
| 48 | F | RAEB-1 | i (17),+9,+13 | i (17) |
| 51 | F | RAEB-2 | +8,-X | +8 |
| 6 | M | RCMD | Normal | del (7q) |
| 55 | M | RCMD | Normal | del (20q) |
| 57 | M | RAEB-2 | Normal | add (16),-17 |
| 59 | F | RAEB-2 | Normal | +9, i (17) |
| 46 | F | RCMD | +8, der (20;21),+del (22q) | Complex |
| 65 | M | RCMD | +8 | +8, +8 |
| 64 | M | RCMD | Complex | NT |
| 2 | F | RAEB-1 | Normal | NT |
| 4 | M | RAEB-2 | Normal | NT |
| 21 | F | RAEB-2 | Normal | NT |
| 47 | M | RAEB-2 | Normal | NT |
| 31 | F | RA | NT | Normal |
| 61 | M | RA | NT | Normal |
| 65 | M | RCMD | NT | del (12q) |
| 40 | M | RAEB-2 | NT | t (6;9) |
| 42 | F | RAEB-2 | NT i | inv (3), del (5q) |
| 43 | M | RAEB-2 | NT | Normal |
| 79 | M | RAEB-2 | NT | Normal |
Table 3.
Results of immunohistochemistry for NPM and sequencing for exon 12 of NPM1
| Patients | At MDS | At AML | ||||
|---|---|---|---|---|---|---|
| Age | Sex | Initial diagnosis | IHC | Sequencing | IHC | Sequencing |
| 63 | M | RA | NPMc+ | Type B | NPMc+ | Type A |
| 60 | F | RAEB1 | NPMc– | Wild | NPMc+ | Type B |
| 43 | M | RAEB2 | NPMc– | Wild | NPMc+ | Type B |
| 57 | M | RAEB2 | NPMc+ | Wild | NT | Type B |
| 29 | M | RAEB2 | NPMc– | Wild | NPMc+ | Type B |
Table 4.
Characteristics of patients with FLT3-ITD at AML transformation
Table 5.
Relationship between FLT3-ITD results at AML transformation and clinical characteristics.
| FLT3-ITD at AML transformation | P value | ||
|---|---|---|---|
| Positive (N=5) | Negative (N=29) | ||
| Time to AML (mon)∗ | 3.6 (3.6–4.4) | 9.2 (4.1–18.2) | 0.232 |
| N of CR achievement (%) | 3 (60.0) | 19 (65.5) | 0.274† |
| Time to CR achievement (mon)∗ | 4.7 (3.7–5.1) | 1.2 (1.1–1.6) | 0.007 |
| Duration of CR (mon)∗‡ | 2.7 (2.4–21.4) | 5.7 (3.9–28.3) | 0.573 |
| Survival rate (%) | 1/5 (20.0) | 6/25 (24.0) | 0.671† |
| Overall survival (mon)∗ | 9.1 (7.1–25.4) | 26.3 (13.1–48.5) | 0.163 |
| Survival after AML (mon)∗ | 5.5 (3.9–7.6) | 9.4 (2.3–30.9) | 0.888 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download