Abstract
Objectives
Osteoclasts (OCs) are bone-resorbing multinucleated cells derived from hematopoietic progenitors of the monocyte-macrophage lineage. OC precursors, such as bone marrow-derived macrophages (BMMs), are formed in the presence of macrophage colony-stimulating factor (M-CSF) and differentiate into OCs in response to M-CSF and receptor activator of nuclear factor κB ligand (RANKL). In this study, we investigated the role of mixed lineage kinases (MLKs)-c-Jun amino-terminal kinase (JNK) pathways in OC formation.
Methods
We performed an OC formation assay and reverse transcription polymerase chain reaction (RT-PCR) analysis.
Results
We first explored the role of JNK on osteoclst formation using mouse bone marrow (BM) culture system. We found that OC formation was impaired when the JNK inhibitor was added either in early or late stage, suggesting the requirement for JNK activation during OC formation. MLKs are serine/threonine kinases that regulate signaling by the JNK. Since the JNK activity is specifically required for osteoclastogenesis, we examined the messenger ribonucleic acid (mRNA) levels of MLKs in BMs, BMMs and OCs by RT-PCR. Among MLKs, the level of MLK3 mRNA expression is highest in BMs, BMMs and OCs. Moreover, we found that the mRNA expression of MLK2 and MLK3 is increased with the differentiation of BMs to BMMs, and is sustained in OCs. Finally we investigated the role of MLK3 in OC differentiation using gene knock-down techniques. The silencing of MLK3 in BMMs partly attenuated RANKL-induced OC differentiation.
Figures and Tables
 | Fig. 1
Effects of JNK inhibitor on osteoclatstogenesis. (A) Schematic design for treatment of JNK inhibitor. (B) Bone marrow cells were cultured with 10 ng/mL M-CSF overnight. Non-adherent cells were harvested and cultured with 30 ng/mL M-CSF in the absence or presence of SP for 3 days. Floating cells were removed and adherent cells were used as BMMs (①). BMMs were cultured with 30 ng/mL M-CSF and 200 ng/mL RANKL in the absence or presence of SP for 4 days (②). Cultured cells were fixed and stained for TRAP. TRAP+ multinucleated cells containing more than three nuclei were counted as osteoclasts. Data represent means ± SD of triplicate samples in a representative experiment. *P < 0.05 versus vehicle. (JNK, c-Jun amino-terminal kinase; M-CSF, macrophage colony-stimulating factor; SP, SP600125 [a JNK inhibitor] BMM, bone marrow-derived macrophage; RANKL, receptor activator of nuclear factor κB ligand; TRAP, tartrate-resistant acid phosphatase; MNC, multinucleated cells) |
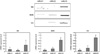 | Fig. 2
The mRNA expression levels of MLKs in BMs, BMMs and OCs. Whole cell lysates were harvested from cultured cells. Total RNA were then isolated from the cells and cDNA templates prepared. The mRNA expression level was determined by RT-PCR using specific primers designed for MLK1, MLK2 and MLK3. Data represent means ± SD of three independent experiments. (mRNA, messenger ribonucleic acid; MLK, mixed lineage kinase; BM, bone marrow; BMM, bone marrow-derived macrophage; OC, Osteoclast; cDNA, complementary deoxyribonucleic acid; RT-PCR, reverse transcription polymerase chain reaction) |
 | Fig. 3
Expression pattern of MLKs in BM and BMM. (A, B) Whole cell lysates were harvested from cultured cells and analyzed by RT-PCR. Data represent means ± SD of three independent experiments. *P < 0.05 versus BM. (N.S., non-significant; MLK, mixed lineage kinase; BM, bone marrow; BMM, bone marrow-derived macrophage; RT-PCR, reverse transcription polymerase chain reaction) |
 | Fig. 4
Expression pattern of MLKs in BMM and OC. (A, B) Whole cell lysates were harvested from cultured cells and analyzed by RT-PCR. Data represent means ± SD of three independent experiments. *P < 0.05 versus BMM. (N.S., non-significant; CRT, calcitonin receptor, MLK, mixed lineage kinase; BMM, bone marrow-derived macrophage; OC, Osteoclast; RT-PCR, reverse transcription polymerase chain reaction) |
 | Fig. 5
Effects of MLK3 knock-down on osteoclatstogenesis. (A, B) BMMs were transfected with non-targeting siRNA or siRNA specific for MLK3. (A) Transfected BMMs were harvested from cultured cells and analyzed by RT-PCR. (B) Transfected BMMs were cultured for 4 days with 30 ng/mL M-CSF and 200 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. TRAP-positive (+) multinucleated cells (MNCs) having more than 3 (n > 3) or 10 (n > 10) nuclei were counted. Data represent means ± SD of triplicate samples in a representative experiment. *P < 0.05 versus siNT. scale bar = 200 µM. (siNT, small interfering non-targeting; MLK, mixed lineage kinase; TRAP, tartrate-resistant acid phosphatase; MNC, multinucleated cells; siRNA, small interfering ribonucleic acid; BMM, bone marrow-derived macrophage; RT-PCR, reverse transcription polymerase chain reaction; M-CSF, macrophage colony-stimulating factor; RANKL, receptor activator of nuclear factor κB ligand; TRAP, tartrate-resistant acid phosphatase) |
References
1. Takahashi N, Akatsu T, Udagawa N, et al. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988. 123:2600–2602.

2. Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999. 20:345–357.

3. Wong BR, Rho J, Arron J, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J Biol Chem. 1997. 272:25190–25194.

4. Yasuda H, Shima N, Nakagawa N, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci U S A. 1998. 95:3597–3602.

5. Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998. 93:165–176.

6. David JP, Sabapathy K, Hoffmann O, Idarraga MH, Wagner EF. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J Cell Sci. 2002. 115:4317–4325.

7. Chang EJ, Ha J, Huang H, et al. The JNK-dependent CaMK pathway restrains the reversion of committed cells during osteoclast differentiation. J Cell Sci. 2008. 121:2555–2564.

8. Otero JE, Dai S, Foglia D, et al. Defective osteoclastogenesis by IKKbeta-null precursors is a result of receptor activator of NF-kappaB ligand (RANKL)-induced JNK-dependent apoptosis and impaired differentiation. J Biol Chem. 2008. 283:24546–24553.

9. Ikeda F, Matsubara T, Tsurukai T, Hata K, Nishimura R, Yoneda T. JNK/c-Jun signaling mediates an anti-apoptotic effect of RANKL in osteoclasts. J Bone Miner Res. 2008. 23:907–914.

10. Yamamoto A, Miyazaki T, Kadono Y, et al. Possible involvement of IkappaB kinase 2 and MKK7 in osteoclastogenesis induced by receptor activator of nuclear factor kappaB ligand. J Bone Miner Res. 2002. 17:612–621.

11. Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002. 3:663–672.

12. Handley ME, Rasaiyaah J, Chain BM, Katz DR. Mixed lineage kinases (MLKs): a role in dendritic cells, inflammation and immunity? Int J Exp Pathol. 2007. 88:111–126.

13. Ciallella JR, Saporito M, Lund S, et al. CEP-11004, an inhibitor of the SAPK/JNK pathway, reduces TNF-alpha release from lipopolysaccharide-treated cells and mice. Eur J Pharmacol. 2005. 515:179–187.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download