Abstract
Polymerase chain reaction (PCR) can detect bacteria more rapidly than conventional plate counting. However DNA-based assays cannot distinguish between viable and dead cells due to persistence of DNA after cells have lost their vitality. Recently, propidium monoazide (PMA) treatment has been introduced. The purpose of this study is to evaluate the applicability of the PMA treatment and real-time PCR method for cell counting in comparison with plate counting and to evaluate the antibacterial efficacy of 2% CHX on E. faecalis using PMA treatment in combination with real-time PCR.
Firstly, to elucidate the relationship between the proportion of viable cells and the real-time PCR signals after PMA treatment, mixtures with different ratios of viable and dead cells were used. Secondly, relative difference of viable cells using PMA treatment in combination with real-time PCR was compared with CFU by plate counting. Lastly, antibacterial efficacy of 2% CHX on E. faecalis was measured using PMA treatment in combination with real-time PCR.
The results were as follows :
Ct value increased with decreasing proportion of viable E. faecalis.
There was correlation between viable cells measured by real-time PCR after PMA treatment and CFU by plate counting until Optical density (OD) value remains under 1.0. However, viable cells measured by real-time PCR after PMA treatment have decreased at 1.5 of OD value while CFU kept increasing.
Relative difference of viable E. faecalis decreased more after longer application of 2% CHX.
Figures and Tables
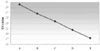 | Figure 1Ct value measured by real-time PCR after serial dilutions of E. faecalis (A. 105 dilution, B. 104 dilution, C. 103 dilution, D. 102 dilution, E. 10 dilution) |
 | Figure 2Effect of PMA on the amplification of different ratios of viable and dead E. faecalis (A. Table showing mixing ratios of viable and dead E. faecalis, B. Ct value of amplified genomic DNA shown as a function of the percentage of viable E. faecalis, C. Relative difference of viable E. faecalis measured by real-time PCR) |
 | Figure 3Correlation between relative difference measured by real-time PCR and log CFU by plate counting (group A. Relative difference of non-PMA treated E. faecalis at each optical density, group B. Relative difference of PMA treated E. faecalis at each optical density, group C. Log CFU by plate counting at each optical density) |
References
1. Williams JM, Trope M, Caplan DJ, Shugars DC. Detection and quantitation of E. faecalis by real-time PCR (qPCR), reverse transcription-PCR (RT-PCR), and cultivation during endodontic treatment. J Endod . 2006. 32:715–721.

2. Young G, Turner S, Davies JK, Sundqvist G, Figdor D. Bacterial DNA persists for extended periods after cell death. J Endod. 2007. 33:1417–1420.

3. Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol. 1993. 59:3513–3515.

4. Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol. 2007. 73:5111–5117.

5. Nocker A, Sossa KE, Camper AK. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J Microbiol Methods. 2007. 70:252–260.

6. Molander A, Reit C, Dahlen G, Kvist T. Microbiological status of root-filled teeth with apical periodontitis. Int Endod J. 1998. 31:1–7.

7. Möller AJ. Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol Tidskr. 1966. 74:1–380.
8. Sundqvist G, Figdor D, Persson S, Sjögren U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1998. 85:86–93.

9. Rôças IN, Jung IY, Lee CY, Siqueira JF Jr. Polymerase chain reaction identification of microorganisms in previously root-filled teeth in a South Korean population. J Endod. 2004. 30:504–508.

10. Haapasalo M, Orstavik D. In vitro infection and disinfection of dentinal tubules. J Dent Res. 1987. 66:1375–1379.
11. Wilson M. Susceptibility of oral bacterial biofilms to antimicrobial agent. J Med Microbiol. 1996. 44:79–87.
12. Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001. 9:34–39.

13. Lima KC, Fava LR, Siqueira JF Jr. Susceptibilities of Enterococcus faecalis biofilms to some antimicrobial medications. J Endod. 2001. 27:616–619.

14. Fouad AF, Kum KY, Clawson ML, Barry J, Abenoja C, Zhu Q, Caimano M, Radolf JD. Molecular characterization of the presence of Eubacterium spp. and Streptococcus spp. in endodontic infections. Oral Microbiol Immunol. 2003. 18:249–255.

15. Abdullah M, Ng YL, Gulabivala K, Moles DR, Spratt DA. Susceptibilities of two Enterococcus faecalis phenotypes to root canal medications. J Endod. 2005. 31:30–36.

16. Portenier I, Waltimo T, Ørstavik D, Haapasalo M. The susceptibility of starved, stationary phase, and growing cells of Enterococcus faecalis to endodontic medicaments. J Endod. 2005. 31:380–386.

17. Zamany A, Spångberg LS. An effective method of inactivating chlorhexidine. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002. 93:617–620.

18. Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006. 67:310–320.

19. Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol. 2006. 72:1997–2004.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download