Abstract
A 3-month-old male cat in the animal facility was presented for investigation of anorexia and occasional vomiting. We collected the specimens from gastroscopic biopsy and stool collection. The gastroscopic biopsy specimens were tested using a rapid urease test, CLO Helicobacter-detection kits. Stool specimens were gathered and evaluated using the commercially available SD Bioline H. pylori Ag kit according to the manufacturer's instructions. Genomic DNAs from gastroscopic biopsy and stool specimens of the cat were extracted and submitted to the consensus PCR to amplify Helicobacter rpoB gene. Then the DNAs from gastroscopic biopsy and stool specimens were conducted a multiplex species-specific PCR to amplify urease B gene for H. heilmannii, H. pylori and H. felis. As the results, the rapid urease test with gastroscopic biopsy was revealed positive reaction. The result of H. pylori Stool Ag assay was one red line, negative for H. pylori. The gastroscopic biopsy and stool specimen were positive reactions by the consensus PCR reaction using the RNA polymerase beta-subunit-coding gene (rpoB) to detect Helicobacter species. By multiplex species-specific PCR with gastroscopic biopsy and stool specimens, no amplification products corresponding to either H. heilmannii or H. pylori were detected, but the specimens tested were positive for H. felis. This case was confirmed as gastroenteric disease induced by H. felis infection. On our knowledge, this is a very rare report about H. felis-induced gastroenteric disease in cat and may provide a valuable data on the study of feline Helicobacter infection.
Gastric bacteria categorized in the genus Helicobacter are Gram negative and spiral shaped; at least 24 species have been reported, and most Helicobacter spp. are suspected or proved gastric or hepatic pathogens [1]. It is likely that several novel Helicobacter species await discovery. Members of this genus are microaerobic, have a fusiform or curved to spiral rod morphology and are motile by flagella that vary in number and location among different species [2]. All known Helicobacters live in human and animal hosts, where colonization occurs primarily in the gastrointestinal tract. Helicobacters colonize the stomachs and intestines of humans and several animal species, such as cats, dogs, ferrets, pigs, cheetahs, and monkeys [3]. In humans, Helicobacter pylori is the major agent of chronic diffuse superficial gastritis, plays a causative role in peptic ulcers, and is considered a cofactor in the development of gastric malignancies. H. pylori has also been found in cats [4], and it can promote gastritis when introduced into specific-pathogen-free cats [5]. The significance of this infection as a cause of gastritis in pet cats is nevertheless unclear. The main gastric Helicobacter species in cats are primarily Helicobacter heilmannii and Helicobacter felis [3]. The prevalence of those two species in cats has been reported to be between 57 and 100% [6]. So far, H. heilmannii has not been reliably cultured in vitro [7]. However, the two organisms can be identified by electron microscopy or by comparison PCR of their 16S rRNA and urease gene sequences [89].
Thus, H. pylori diagnosis is based on detecting urease. Several methods have been proposed and used to diagnose H. pylori infection. Increasing interest has been directed toward non-invasive tests, compared to endoscopy-based invasive methods (histology and rapid urease test), as non-invasive methods do not require endoscopic assessment [10]. The 13C-urea breath test (UBT) is the most recommended non-invasive test for detecting H. pylori infection and has high sensitivity and specificity [11]. However, the UBT cannot be applied to animals due to its high cost and the requirement for expensive analytical instruments [12]. Thus, many researchers have used polymerase chain reaction (PCR) assays to monitor the infection in stools without biopsy or sacrifice of animals [1314]. However, PCR assay need time-consuming and high techniques and high-cost laboratory instrument like as Thermal Cycler [15]. Furthermore, stool samples remain the most difficult specimens for DNA extraction and amplification [16]. Recently, several companies have been released H. pylori stool antigen (HpSA) test kits. HpSA tests are non-invasive diagnostic modules for H. pylori infection with human patient's stool samples [171819]. However, there was little information about the usefulness of HpSA test on the H. felis, which is a major Helicobacter species in cats.
In this study, we detected a gastric Helicobacter species-infected cat with gastric disease by a Consensus PCR analysis and the rapid urease test. Thereafter, Helicobacter felis was identified by a species-specific PCR.
A 3-month-old male cat was obtained from the Animal Facilities of the Center for Animal Resources Development, Wonkwang University, Korea. The animal experiments in this study were conducted according to the ethical procedures of Wonkwang University IACUC. The cat was presented for investigation of anorexia and occasional vomiting. Abdominal radiography and sonography did not demonstrate any abnormal lesions. The possibility of feline leukemia, feline immunodeficiency virus and heartworm was ruled out from the results of rapid test kits (Bionote, Suwon, Korea).
Fasting cat was anesthetized with diazepam (0.2 mg/kg of body weight) and ketamine (3 to 5 mg/kg given until effective); the cat was intubated, and anesthesia was maintained with halothane in oxygen. During gastroscopy the macroscopic appearance of the mucosa was recorded and biopsy samples were taken. Endoscope and biopsy forceps were disinfected with 4% Sekusept Plus solution (Henkel, Muttenz, Switzerland) for 30 min and thoroughly flushed with tap water. Gastroscopic biopsy specimens were submitted to rapid urease test (RUT) and PCR assay.
Stool specimens collected in sterile screw-capped containers were transported and stored at room temperature. Specimens were processed for PCR analysis within 24 h of being collected. Each collection swabs were put into 2 mL of 0.1 M PBS buffer and vortexed and discarded, and the PBS was submitted to extract genomic DNAs for PCR assay.
The gastroscopic biopsy specimens were minced and applied to confirm infection using a rapid urease test, CLO Helicobacter-detection kits (Asan Pharm Co., Ltd., Seoul, Korea) by incubating the biopsy specimens at 35℃ for 24 hours to examine urease activity. The reaction (color change) was determined as bright yellow (negative), thick yellow false (partially) positive, or thick (dark) red for positive.
Stool specimens were gathered and evaluated using the commercially available SD Bioline H. pylori Ag kit according to the manufacturer's instructions. Specimens (250 mg) were incubated with diluents solution at room temperature for 30 min and then 100 µL was placed on the Helicobacter Ag examination device. The test results were checked about 15 min later. One red line indicated negative, and double red line indicated Helicobacter positive result.
Gastroscopic biopsy tissues was homogenized and resuspended in PBS, and DNA was extracted from the homogenates. Briefly, genomic DNA was isolated using an AccuPrep Genomic DNA Extraction kit (Bioneer Corp., Daejeon, Korea), according to the manufacturer's instructions. DNA was eluted in Tris-EDTA buffer (pH 8.0), and an aliquot was used for PCR amplification. All DNA samples were stored at -20℃ until the PCR assays were performed.
Genomic DNAs from stool specimens of the cat were extracted using the AccuPrep Stool DNA Extraction Kit (Bioneer) according to the manufacturer's instructions. AccuPrep Stool DNA Extraction Kit is designed for the rapid, convenient extraction of DNA from fresh or frozen stool, or other samples containing large amounts of material that can inhibit PCR. The kit uses a glass filter, fixed in a column tube that can efficiently bind DNA in the presence of chaotropic salts. Using the spin-column method, contaminants and enzyme inhibitors (such as heparin, bilirubin bile salts, porphyrin) are eliminated and high-purity DNA is obtained, ready for use in a variety of applications [15]. After washing steps which remove proteins and salt, high-purity DNA is finally eluted using a low-concentration elution buffer. It provides 2~5 µg DNA yields from 100 mg stool [15].
A set of primers (HF, 5'-ACTTTAAACGCATGAAG ATAT-3'; and HR, 5'-ATATTTTGACCTTCTGGGGT-3') was used to amplify rpoB DNA (458 bp) encompassing the Rifr region [15]. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (Maxime PCR PreMix; iNtRON Biotechnology, Seongnam, Korea) containing 1 U of Taq DNA polymerase, 250 µM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and the gel loading dye. The volume was adjusted with distilled water to 20 µL. The reaction mixture was subjected to 40 amplification cycles (5 min at 95℃, 30 s at 94℃, 30 s at 52℃, 45 s at 72℃, and 5 min at 75℃) followed by a 5-min extension at 72℃ (model 9600 Thermocycler; Perkin-Elmer Cetus, Waltham, MA). The PCR products were electrophoresed on a 1.2% (wt/vol) agarose gel. Positive samples were further analyzed using species-specific PCR and DNA sequencing to identify the individual Helicobacter species.
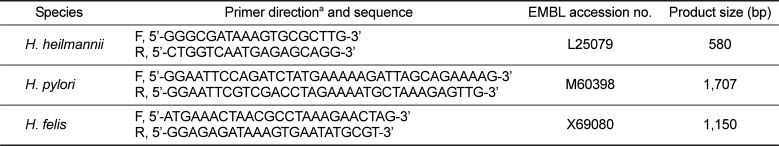
For multiplex species-specific PCR, primers were derived from the urease B sequence of each Helicobacter species. Table 1 showed sequences, EMBL accession numbers, and product lengths [14]. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (Maxime PCR PreMix; iNtRON Biotechnology) containing 1 U of Taq DNA polymerase, 250 µM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and the gel loading dye. PCRs were performed as follows: one cycle of 94℃ for 3 min, 57℃ for 2 min, and 72℃ for 3 min followed by 31 cycles of 94℃ for 30 s, 57℃ for 30 s, and 72℃ for 1 min and a final extension at 72℃ for 5 min. PCR products were analyzed by electrophoresis on a 1.2% agarose gel. Amplified products for H. heilmannii (580 bp), H. pylori (1,707 bp) and H. felis (1,150 bp) were identified.
The gastroscopic biopsy specimens were tested using a rapid urease test, CLO Helicobacter-detection kits. The reaction (color change) was determined as bright yellow (negative), thick yellow false (partially) positive, or thick (dark) red for positive. The result was thick red colored, positive result (Figure 1)
We collected stool specimens, which contained less water. We incubated the stool specimens in diluent solution for 30 min with vortexing every 10 min. Then, we placed 100 µL of sample on the SD Bioline H. pylori Stool Ag kit detection device. One red line indicated negative, and double red line indicated Helicobacter positive result. The result was one red line, negative for H. pylori (Figure 2).
The gastroscopic biopsy tissues were homogenized and resuspended in PBS, and DNAs were extracted from the homogenates of gastric tissues. Genomic DNAs from stool specimens of the cat were extracted using the AccuPrep Stool DNA Extraction Kit. A consensus PCR analysis method using the RNA polymerase beta-subunit-coding gene (rpoB) was employed to detect Helicobacter species. Amplification of rpoB DNAs from consensus PCR products was electrophoresed on 1.2% agarose gel. The detection limit of the consensus PCR is 0.1 pg of Helicobacter DNA in stool [15]. As the results, the gastroscopic biopsy and stool specimen were positive reactions by the consensus PCR reaction (Figure 3).
Multiplex species-specific PCR with gastroscopic biopsy and stool specimens was conducted with the primers using urease B gene. No amplification products corresponding to either H. heilmannii or H. pylori were detected, but the specimens tested were positive for H. felis (Figure 4).
PCR is a specific method to distinguish between different Helicobacter spp. It is also sensitive for the diagnosis of H. pylori in humans and cats [320]. Cats can be infected with different Helicobacter species, and mixed infections occur [21]. Furthermore, H. pylori has been detected in several feline stomachs [4], but the public health implication of this observation is unclear [22]. While some human epidemiological studies showed contact with cats to be a risk factor in the acquisition of H. pylori [23], other reports could not confirm this [24]. H. felis is characterized by the presence of periplasmic fibrils [21], while H. heilmannii is not. Species identification based upon such criteria must be made cautiously, since the stability of this in vivo characteristic has not been established. Recently, the in vitro loss of periplasmic fibrils by H. felis was described [25]. Unfortunately, H. heilmannii cannot be grown in vivo [9].
The current gold standard for diagnosing H. pylori infection is endoscopic biopsy of gastric tissue for the rapid urease test, histology, and culture. However, such an invasive procedure has major disadvantages of anesthesia, discomfort, and the possibility for ethical problems [26]. But, noninvasive tests are easy to perform and do not produce significant discomfort and allow a patient to avoid the discomfort and risk of invasive endoscopy. These include serological antibody testing for H. pylori, the urea breath test, and the HpSA test [26].
Among several diagnostic tests, the HpSA test and PCR with the stool specimens for diagnosing H. felis infection may offer a useful non-invasive method without sacrificing animals during an in vivo study or veterinary clinic. In this study, HpSA did not detected H. felis in stool specimen. On the other hand, RUT and PCR assay with the gastric and stool specimens could detected successfully H. felis. Despite all the above observation on performance of antigen detection H. feslis in stool, it has certain disadvantage: antigen excretion may vary over the time period and antigen may degrade while passing through intestine. Further, use of N-acetylcysteine like mucolytic agent may decrease the accuracy of the diagnosis [27]. Cut off titer, though difficult to decide but crucial to reach the conclusion by using antigen detection technique [28]. PCR analysis also has been reported several limitations [15]. There is no single gold standard among the diagnostic tests for Helicobacter infection but all of the tests have their pitfalls and limitations [29]. Among noninvasive methods, stool specimens are easy to obtain and consequently of high potential interest for the development of a direct method of Helicobacter species detection [2630]. PCR was successfully used to detect the bacterium in stools [2630]. Indeed stool samples remain the most difficult specimens for DNA extraction and amplification [16]. The difficulty has been associated with the DNA polymerase inhibitors present in stools, identified as complex polysaccharides [16]. In this study, we used the spin-column method and it eliminated effectively contaminants and enzyme inhibitors (such as heparin, bilirubin bile salts, porphyrin) and high-purity DNA is obtained. It provides 2~5 µg DNA yields from 100 mg stool [30]. We suggest that PCR assay could be successfully used to detect the H. felis in stools.
In conclusion, this case was confirmed as gastroenteric disease induced by H. felis infection. On our knowledge, this is a very rare report about H. felis-infected cat in the laboratory animal facility and may provide a valuable data on the study of feline Helicobacter infection.
Acknowledgments
This study was supported by the research fund of Wonkwang University in 2016. We wish to appreciate Gi-Wook Oh, research assistants of Center for Animal Resources Development, Wonkwang University, for carrying out the technical support.
References
1. Fox JG. The non-H pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut. 2002; 50(2):273–283. PMID: 11788573.

2. Vandamme P, Falsen E, Pot B, Kersters K, De Ley J. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J Clin Microbiol. 1990; 28(5):1016–1020. PMID: 2191002.

3. Kim O. Helicobacter - An emerging new zoonotic pathogen. In : Lorenzo-Morales J, editor. Zoonosis. Rijeka, Croatia: InTec Press;2012. p. 89–100.
4. Handt LK, Fox JG, Dewhirst FE, Fraser GJ, Paster BJ, Yan LL, Rozmiarek H, Rufo R, Stalis IH. Helicobacter pylori isolated from the domestic cat: public health implications. Infect Immun. 1994; 62(6):2367–2374. PMID: 8188360.

5. Fox JG, Batchelder M, Marini RP, Yan L, Handt LK, Li X, Shames B, Hayward A, Campbell J, Murphy JC. Helicobacter pylori-induced gastritis in the domestic cat. Infect Immun. 1995; 63(7):2674–2681. PMID: 7790084.

6. Geyer C, Colbatzky F, Lechner J, Hermanns W. Occurrence of spiral-shaped bacteria in gastric biopsies of dogs and cats. Vet Rec. 1993; 133(1):18–19. PMID: 8362485.

7. Andersen LP, Nørgaard A, Holck S, Blom J, Elsborg L. Isolation of a "Helicobacter heilmanii"-like organism from the human stomach. Eur J Clin Microbiol Infect Dis. 1996; 15(1):95–96. PMID: 8641315.

8. Solnick JV, O'Rourke J, Lee A, Paster BJ, Dewhirst FE, Tompkins LS. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993; 168(2):379–385. PMID: 8335974.

9. Solnick JV, O'Rourke J, Lee A, Tompkins LS. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect Immun. 1994; 62(5):1631–1638. PMID: 8168924.

10. Mégraud F. The most important diagnostic modalities for Helicobacter pylori, now and in the future. Eur J Gastroenterol Hepatol. 2012; 9(Suppl 1):S13–S15. PMID: 22498901.

11. Leal YA, Flores LL, Fuentes-Pananá EM, Cedillo-Rivera R, Torres J. 13C-urea breath test for the diagnosis of Helicobacter pylori infection in children: a systematic review and meta-analysis. Helicobacter. 2011; 16(4):327–337. PMID: 21762274.

12. Nyan DC, Welch AR, Dubois A, Coleman WG Jr. Development of a noninvasive method for detecting and monitoring the time course of Helicobacter pylori infection. Infect Immun. 2004; 72(9):5358–5364. PMID: 15322033.
13. Santos AM, Lopes T, Oleastro M, Chaves P, Cordeiro R, Ferreira M, Pereira T, Machado J, Guerreiro AS. Role of 13C-urea breath test in experimental model of Helicobacter pylori infection in mice. Helicobacter. 2011; 16(4):320–326. PMID: 21762273.

14. Neiger R, Dieterich C, Burnens A, Waldvogel A, Corthésy-Theulaz I, Halter F, Lauterburg B, Schmassmann A. Detection and prevalence of Helicobacter infection in pet cats. J Clin Microbiol. 1998; 36(3):634–637. PMID: 9508286.
15. Kim S, Cho S, Kim O. Detection and identification of secreting Helicobacter species from cats. Lab Anim Res. 2006; 22(3):243–247.
16. Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, Vidal R, Cabrita J, Mégraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997; 35(4):995–998. PMID: 9157172.

17. Moon DI, Shin EH, Oh HG, Oh JS, Hong S, Chung Y, Kim O. Usefulness of a Helicobacter pylori stool antigen test for diagnosing H. pylori infected C57BL/6 mice. Lab Anim Res. 2013; 29(1):27–32. PMID: 23573105.
18. Shimoyama T. Stool antigen tests for the management of Helicobacter pylori infection. World J Gastroenterol. 2013; 19(45):8188–8191. PMID: 24363508.
19. Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: what should be the gold standard? World J Gastroenterol. 2014; 20(36):12847–12859. PMID: 25278682.
20. Mapstone NP, Lynch DA, Lewis FA, Axon AT, Tompkins DS, Dixon MF, Quirke P. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. J Clin Pathol. 1993; 46(6):540–543. PMID: 8331177.

21. Lee A, Hazell SL, O'Rourke J, Kouprach S. Isolation of a spiral-shaped bacterium from the cat stomach. Infect Immun. 1988; 56(11):2843–2850. PMID: 3169989.

22. Handt LK, Fox JG, Stalis IH, Rufo R, Lee G, Linn J, Li X, Kleanthous H. Characterization of feline Helicobacter pylori strains and associated gastritis in a colony of domestic cats. J Clin Microbiol. 1995; 33(9):2280–2289. PMID: 7494015.

23. Morris A, Nicholson G, Lloyd G, Haines D, Rogers A, Taylor D. Seroepidemiology of Campylobacter pyloridis. N Z Med J. 1986; 99(809):657–659. PMID: 3463897.
24. Webb PM, Knight T, Elder JB, Newell DG, Forman D. Is Helicobacter pylori transmitted from cats to humans? Helicobacter. 1996; 1(2):79–81. PMID: 9398882.

25. Eaton KA, Dewhirst FE, Paster BJ, Tzellas N, Coleman BE, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996; 34(12):3165–3170. PMID: 8940465.

26. Hoshina S, Kahn SM, Jiang W, Green PH, Neu HC, Chin N, Morotomi M, LoGerfo P, Weinstein IB. Direct detection and amplification of Helicobacter pylori ribosomal 16S gene segments from gastric endoscopic biopsies. Diagn Microbiol Infect Dis. 1990; 13(6):473–479. PMID: 1703940.

27. Demirturk L, Yazgan Y, Tarcin O, Ozel M, Diler M, Oncul O, Yildirim S. Does N-acetyl cystein affect the sensitivity and specificity of Helicobacter pylori stool antigen test? Helicobacter. 2003; 8(2):120–123. PMID: 12662379.
28. Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ. European Helicobacter Study Group. Management of Helicobacter pylori infection--the Maastricht IV/Florence Consensus Report. Gut. 2012; 61(5):646–664. PMID: 22491499.
29. Rautelin H, Lehours P, Megraud F. Diagnosis of Helicobacter pylori infection. Helicobacter. 2003; 8(Suppl 1):13–20. PMID: 14617213.

30. Lee H, Park Y, Kim O. Prevalence of Helicobacter Species in Feces of Dogs using Polymerase Chain Reaction Analysis. Lab Anim Res. 2007; 23(3):339–344.
Figure 1
The gastroscopic biopsy specimens of the cat were minced and applied to confirm infection using a rapid urease test, CLO Helicobacter-detection kits. The result was thick red colored, positive reaction for H. pylori.
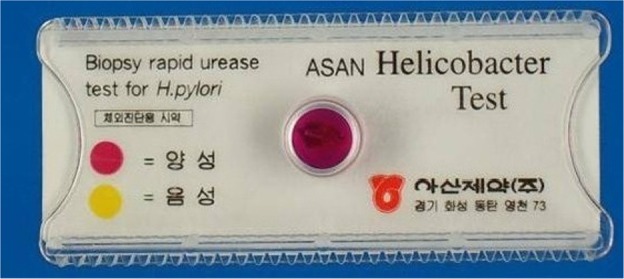
Figure 2
Stool specimens of the cat were gathered and evaluated using the commercially available SD Bioline H. pylori Ag kit. The result was one red line, negative reaction for H. pylori.
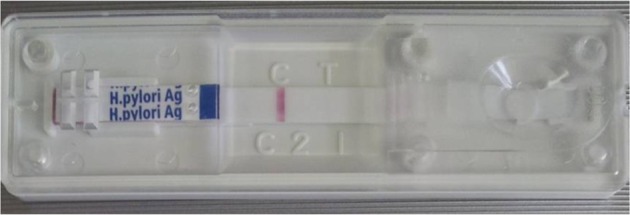
Figure 3
Amplification of Helicobacter rpoB DNAs from consensus PCR was identified on a 1.2% agarose gel electrophoresis. M: Size marker, N: Distilled water, G: Gastroscopic biopsy, S: Stool specimen.
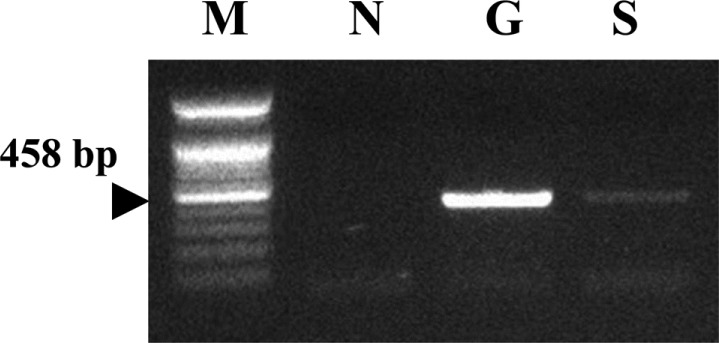
Figure 4
Results of the multiplex species-specific PCR to amplify urease B gene for H. heilmannii, H. pylori and H. felis. M: Size marker, N: Distilled water, G: Gastroscopic biopsy, S: Stool specimen.
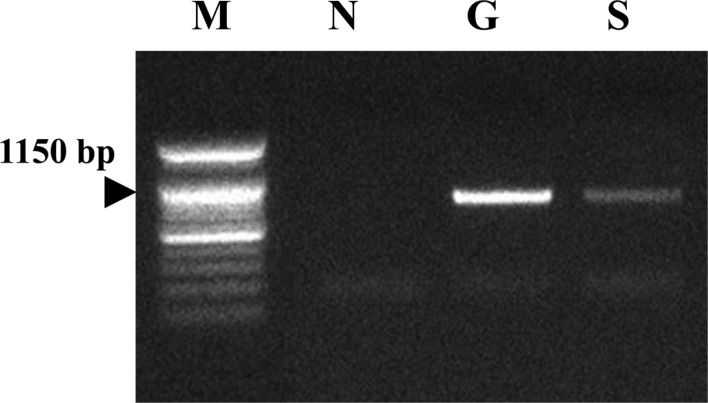
Table 1
Primers used for H. heilmannii, H. pylori, and H. felis urease B gene amplification




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download