Abstract
Pig pancreas may be a therapeutic resource for human diabetic patients. However, this potential is hindered by a lack of knowledge of the molecular events of pig pancreas development. In this study, the embryonic day 60, neonate and 6-month protein profiles of pig pancreas were ascertained at using two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-time of flight mass spectrometry. Twenty four proteins were differentially expressed during pig pancreas development. Among them, 12 spots increased and 7 spots decreased according to development. The expression of 5 protein were highest at birth. Expression of digestive enzymes including trypsin, pancreatic triacylglycerol lipase and pancreatic alpha-amylase was elevated in adults, whereas chymotrypsins were highly expressed in neonates. Proteins that were abundantly expressed during gestation were alpha-1-antitrypsin, alpha-fetoprotein and transferrins. Taken together, we found out that several proteins were significantly up- or down- regulated from pig pancreas based on developmental stage. This study will provide basis for understanding development of pig pancreas.
The pancreas is a complex gland having both exocrine and endocrine activities. The exocrine pancreas in the mature animal synthesizes and stores digestive enzymes to release after ingestion of a meal that induces neural and hormonal stimulation [1]. The endocrine pancreas contains five major cell types, which produce insulin (B cells), glucagon (A cells), somatostatin (D cells), pancreatic polypeptide (PP cells) and ghrelin, which is a novel growth hormone-releasing peptide (ghrelin cells). B cells may also express islet amyloid polypeptide (IAPP), another member of the insulin family of proteins, insulin-like growth factor-II (IGF-II) and pituitary adenylate cyclase activating polypeptide (PACAP) [2].
The pancreas releases these various enzymes and hormones to digest foods and maintain glucose homeostasis. If pancreas is damaged, the aftermath can be serious. Two examples are diabetes mellitus and pancreatic adenocarcinoma. Diabetes mellitus is a chronic metabolic disorder resulting from defects in insulin secretion or function. The global prevalence of diabetes was estimated as 2.8% in 2000 and is predicted to grow to 4.4% by 2030. The total number of diabetics is projected to rise from 171 million in 2000 to 366 million in 2030 due to population growth, aging, urbanization and increasing prevalence of obesity and physical inactivity [3]. Pancreatic adenocarcinoma is the fourth leading cause of cancer deaths in the United States and its prognosis is usually dismal [1].
Preventive and effective treatments of pancreas-related diseases including diabetes mellitus, pancreatitis and pancreatic cancer require knowledge of the biology and molecular characteristics of the pancreas. Better understanding of pancreatic development and regeneration will provide valuable information about the identification of molecular markers involved in the pathogenesis of pancreatic cancer. As well, marker identification can be useful for islet cell transplantation, which is being explored as a treatment for diabetes.
Few proteomic studies have been done using human pancreatic tissues to elucidate the mechanisms of diabetes mellitus and pancreatic cancers because of difficulty in preparing human samples [4,5,6,7]. Instead, in vivo experiments have mainly used animal models. Several studies have contributed to the understanding of the molecular network governing developing pancreas in mouse and rat models [8,9].
The pig has become an attractive subject of study because of its potential use for xenotransplantation [10]. Nonhuman primate, such as apes and monkeys, more closely resemble humans anatomically and physiologically than do pigs. Nonetheless, xenotransplantation using nonhuman primates as xenograft donors is unlikely primarily because of the infectious risks to human patients and their contacts. Pigs produce large litters (up to 10 littermates), have a short gestation time (4 months), are somewhat similar to humans anatomically and physiologically, are available in large numbers (an estimated 90 million pigs are raised for human consumption yearly in the United States) and have a long history of providing medicinal products (skin, insulin, cardiac prostheses, clotting factors) for humans. For these reasons, the pig has become the most likely candidate for consideration as an organ donor [11,12,13,14]. Hindering this potential, the development of pig pancreas is not well understood.
In this study we investigated changes in the proteomic profile during pig pancreas development in embryonic day 60 (E60), neonatal and 6-month-old pigs using two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS).
Pancreas of E60 (n=4), neonate (n=3), 6-month-old male pigs (n=3) were removed immediately after sacrifice and stored in liquid nitrogen until used. Frozen pancreas tissues were homogenized in 400 µl lysis buffer (7 M urea, 2M thiourea, 2% w/v CHAPS, 2% Pharmalyte pH3-10, 100 mM DTE). After centrifugation (50,000 rpm at 4℃ for 1 h) the supernatant was extracted and complete protease inhibitor cocktail (Roche Applied Science, Basel,, Germany), DNase I (2.5 mg/mL; Roche Applied Science) and RNase (2.5mg/mL; Roche Applied Science) were added. The protein concentration was measured by spectrophotometry using Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA) based on the Bradford method. Because urea, thiourea, CHAPS and other constituents of the sample/lysis buffer can interfere with the protein estimation, the standard curve was generated using bovine serum albumin (BSA) at concentrations of 0, 2, 5, 7 and 10 mg/mL. In this process, pancreas sample was made from a pool of each of the 3~4 animals in each group, and each pooled sample was run 3 times.
2-DE was performed as previously described [15]. One milligram quantities of protein were diluted in the sample buffer with 2% IPG buffer up to 450 µL. Samples were added to pH 3-10, nonlinear, 24 cm IPG strips (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and covered with mineral oil. After rehydration for 16 h at 20℃, isoelectric focusing was carried out in eight steps under step-n-hold mode as follows: (S1) 30 V, 2 h; (S2) 100 V, 2 h; (S3) 200 V, 1 h; (S4) 500 V, 1 h; (S5) 1,000 V, 1 h; (S6) gradient, 8,000 V, 1 h; (S7) gradient, 100,000 V, 1 h and (S8) 20,000 VHrs. The total voltage applied was 129,960 Vh (IPGphore Isoelectric Focusing Unit: Amersham Pharmacia Biotech). The IPG strips were equilibrated in buffer containing 6M urea, 20% glycerol, 1.5 M Tris-HCl, pH 8.8, 2.5% acrylamide and Tributyl phosphine (Fluka, Buchs SG, Switzerland). After equilibration, SDS-PAGE was performed by 8-18% gradient gel without stacking gel via the Ettan Dalt system (Amersham Phamacia Biotech). 2-DE was carried out overnight at 4W per gel at 20℃. After finishing 2-DE, the proteins on gels were fixed with protein fixing solution (40% methanol, 5% phosphoric acid) for 2 h and then stained using Coomassie G-250 (Bio-Rad) for 3 h. To remove extra dye in gels, the gels were washed with distilled water.
The stained gels were scanned using a GS-800 calibrated densitometer (Bio-Rad) and spot analysis was performed with the ImageMaster™ 2D platinum software version 5.0 (Amersham Pharmacia Biotech). The digitalized 2-DE gel images were compared by matching using Image master 5.0 (Amersham Biosciences, Piscataway, NJ, USA).
Spots of interest on the 2-DE gel were excised, cut into small pieces and digested with 12.5 ng/µL trypsin (Promega, Madison, WI, USA) in 50 mM ammonium bicarbonate (pH 8.0), as previously described [16]. The preparation procedures for MALDI-TOF MS used reversed-phase columns as previously described [17]. The tryptic peptides were passed through a POROS 50 R2 column (Applied Biosystems, Forster City, CA, USA). After washing the column with 70% acetonitrile in 5% formic acid, 100% acetonitrile and 5% formic acid, peptides were loaded into column and washed with 5% formic acid. The peptides were eluted with 10 mg/mL a-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich, St. Louis, MO, USA) containing solution and elutes were dropped on a MALDI sample plate [18]. Mass analyses were performed using a Voyger-DE PRO MALDI-TOF MS apparatus (Applied Biosystems, Franklin Lakes, NJ, USA), equipped with a 337 nm nitrogen laser. This machine was operated at an accelerating voltage of 20 K, positive ion reflection mode, voltage grid of 74.5%, guide wire voltage of 0% and a delay time of 110 ns. The spectra were externally calibrated using a standard peptide mixture containing angiotensin I, 1,296.68 [M+H]; Glu-Hibrinopeptide B, 1,570.67 [M+H]; ACTH (clip18-39), 2,465.19[M+H]). The proteins were identified by Swiss-Prot and MSDB protein database using MS-FIT (Protein Prospector; UCSF, San Francisco, CA, USA). All the cases of search were analyzed with a 100 ppm mass tolerance and 1 miss cleavage.
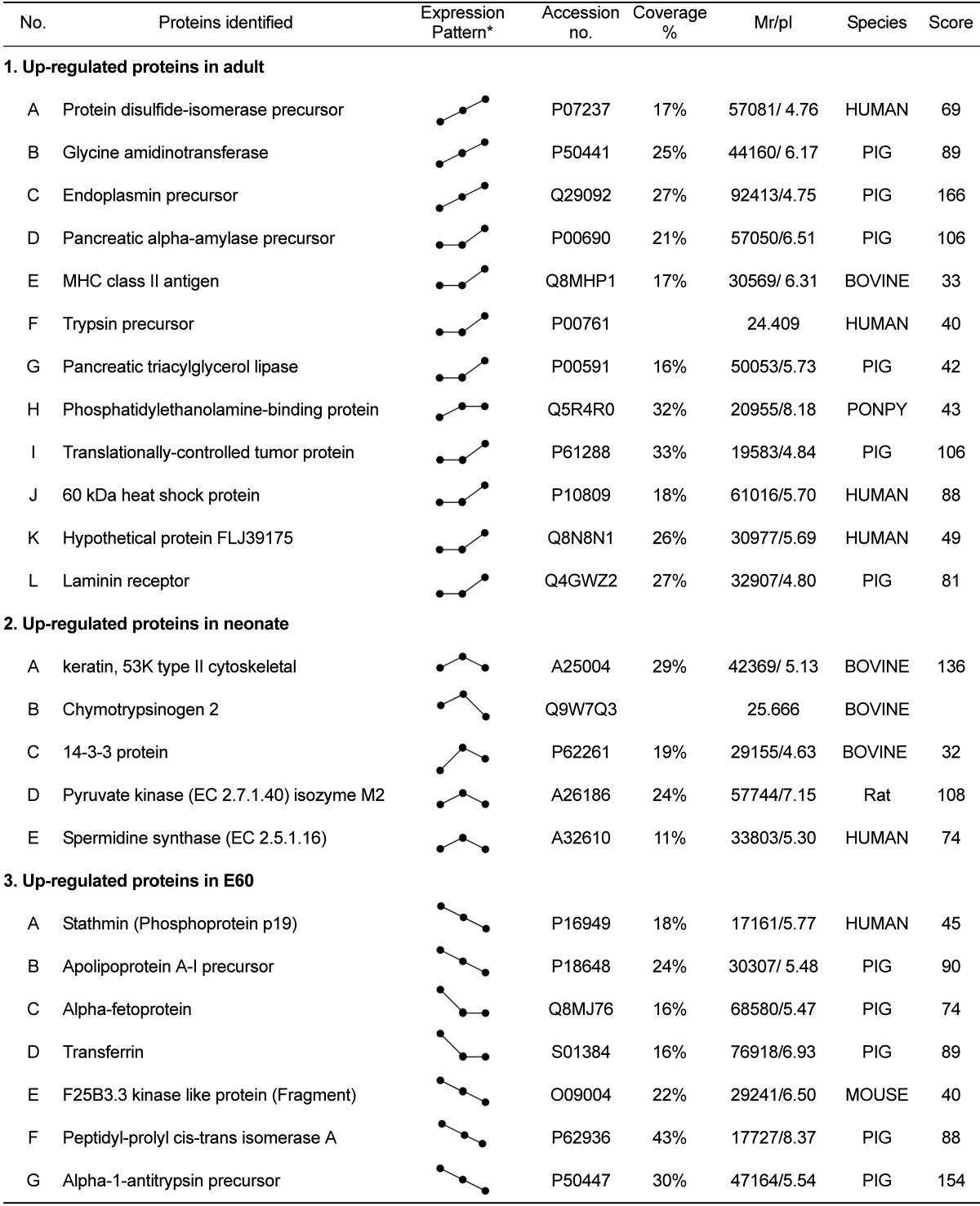
To analyze the changes in the proteome profile during pig pancreas development, the pancreas of E60, neonate, and 6 month-old pigs were used. Pancreatic tissue was processed for 2-D electrophoresis to isolate the candidate proteins that were differentially expressed during development. The change of protein expression was detected in each of the Coomassie blue stained gels using computer-assisted image analysis. At each respective developmental stage, a total of 467, 402 and 557 spots were detected from each gel (Figure 1). Among them, 24 proteins showed marked changes in expression. Twelve spots increased, while 7 spots decreased. The expression level of 5 spots was highest at birth. Twenty four differentially expressed proteins were identified by MALDI-TOF MS and MASCOT (Table 1). Changes in protein expression patterns were visualized in gels.
The amount of protein disulfide-isomerase precursor, endoplasmic precursor, glycine aminotransferase and heat shock protein increased gradually during the development from fetus to adult pig (Figure 2). Their increase suggests that gradual increase in body mass and food uptake of animal results in higher protein content of these proteins for making the necessary quantities of digestive enzymes and hormones. In addition, digestive enzyme precursors, such as pancreatic alpha-amylase precursor, trypsin precursor and pancreatic triacylglycerol lipase, were stage-specific proteins dominantly expressed in adult pig pancreas. Immune-mediated MHC class II antigen was expressed in large quantities in the adult pancreas.
Expression of 5 proteins were up-regulated at the time of birth: chymotrypsinogen 2, pyruvate kinase isozyme M2, spermidine synthase and 14-3-3 protein (Figure 3). Among them, the change in expression pattern of chymotrypsinogen 2 was especially pronounced. However, although chymotrypsinogen 2 was rarely expressed in adult pig pancreas, it occupied at least 20% of entire pancreas proteins in fetus and neonate, especially neonates. The specific activity of chymotrypsin increased until birth and then declined precipitously, consistent with prior reports [19,20].The activities of other zymogens (amylase, lipase and elastase) were low in the fetus and steadily increased after birth.
Expression of 7 spots was significantly increased in fetal pigs compared to neonate and adult stages (Figure 4). Apolipoprotein precursor, alpha-fetoprotein, transferring and alpha-1-antitrypsin precursor were strong markers of developing pancreas. They were difficult to detect in the developed pancreas.
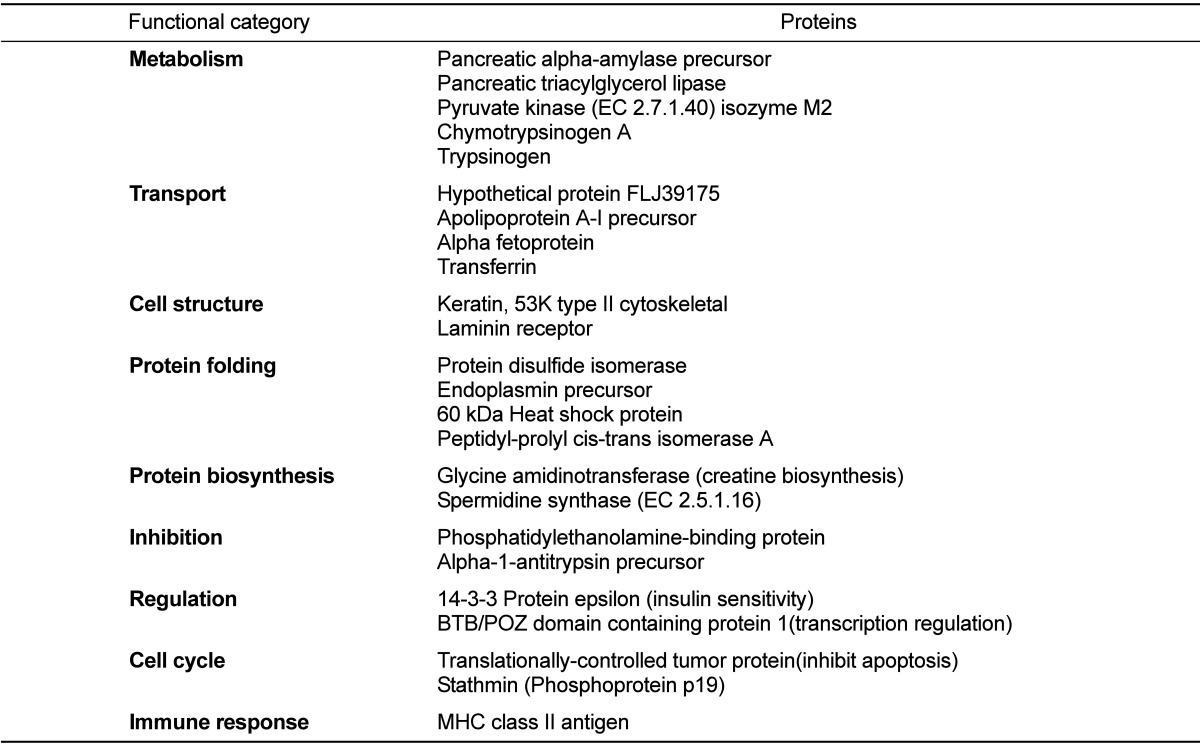
The 24 identified proteins were classified by their functions based on a search at http://harvester.embl.de (Table 2). The functions included glucose or lipid metabolism (n=5), transport of ions like Fe and Ni, or lipid (n=4), maintenance of intracellular structure and cell-to-cell adhesion (n=2), protein folding (n=4), protein synthesis (n=2), anti-proteases (n=2), regulators of signal transduction or insulin sensitivity (n=2), inhibit of apoptosis and induce proliferation (n=2) and immune system related (n=1).
Pancreas obtained prior to birth (E60), at the end of pregnancy (neonate) and at adulthood (6 months of age) were analyzed to reveal proteins that were differentially expressed during pig pancreas development. Using 2D proteomics, proteins were resolved and were identified using MALDI-TOF and MASCOT database. We identified 462, 402 and 557 proteins at E60, neonate and 6-month-old adult pig, respectively. Among those identified proteins, 24 proteins showed pronounced differences in protein expression level at each developmental stage. The fetus and the neonatal pig pancreas had similar protein expression patterns on 2-DE gels, while the adult pig pancreas had a significantly different pattern. In the adult pig, digestive enzymes, such as pancreatic alpha-amylase precursor, trypsin precursor, pancreatic triacylglycerol lipase, protein disulfide-isomerase precursor, endoplasmin precursor and glycine amidinotransferase, which are associated with protein synthesis and formation of tertiary structure, consisted most of the dramatically up-regulated proteins on the 2-DE map.
Comparing protein expression levels in the E60 and the neonatal pig (Figure 1), proteins in the fetal pig were more acidic and high in molecular weights. These included stathmin, apolipoprotein A-I precursor, alpha-fetoprotein, transferrin, peptidyl-prolyl cis-trans isomerase A (cyclophilin A) and alpha-1-antitrypsin precursor. These proteins gradually decreased during pancreas development. Stathmin is the tubulin-binding protein but a neuroproteomic search identified it as a novel Cdk5 substrate. Stathmin is phosphorylated by Cdk5 [21] and is involved in cancer development [22,23]. Stathmin gene overexpression is important in the maintenance of the malignant phenotype in tumor cells, and the blocking efficacy and tumor specificity of this target has been addressed in clinical trials [24]. Small interfering RNA-induced repression of stathmin decreases cell proliferation, viability and clonogenicity in mutant p53 tumor cell lines [25]. Although the relationship between stathmin and cell development is unclear, since stathmin is associated with aberrant cell proliferation, it is expected that stathmin contributes to cell development in some manner.
Apolipoprotein A-I (apoA-I) is the major lipoprotein component of high-density lipoprotein, and plays an important role in reverse cholesterol transport [26]. It also is an anti-inflammatory and anti-atherosclerosis molecule and functions as a potent inhibitor of dendritic cell differentiation and maturation [27]. Apo A-I has two major sites of synthesis: the intestine and the liver. Apo A-I is expressed in the pancreas and 12% of the liver [28]. Presently, its expression decreased gradually according to the developmental stage, which may be due to lack of cell type specification during early development because liver and pancreas have the same developmental origin. In addition, the liver controls lipid metabolism and repression of inflammatory response, which may contribute to the development of the pancreas (although there is scant evidence in support of this).
The liver and yolk sac of fetuses the principle sites of alpha-fetoprotein (AFP) synthesis in ontogenesis, and the dynamics of AFP in ontogenesis from the early embryonic period through mid-pregnancy to pregnancy termination to AFP shut-down in early postnatal period [29]. AFP plays an important role in regulation of immune system, which is associated with repression on the immune system, such as decrease in natural killer cell activity [30], induction of T suppressor cells [31,32], repression of mitogenic response of PHA and Con A [33,34,35,36], restriction on T cell differentiation to Ia determinants [37], decrease in phagocytic activity of macrophage [38,39] and decrease in macrophage Ia expression level [40,41,42].
Transferrin, a 76-80 kDa glycoprotein, is responsible for the transport of iron to cells within both the fetal and maternal systems [43]. It is not only the transporter of iron but also has activities of growth factor and repressing reactive oxygen species production [44]. The concentration of AFP, alpha 1-antitrypsin and transferrin in serum increases with age until 40-50 days of gestation and then decreases progressively at birth in pigs [45].
Cyclophilin A (CyP-A) is a soluble cytoplasmic immunophilin involved in T cell differentiation and proliferation. A role for CyP-A in neuronal differentiation has been implied [46]. Alpha-1-antitrypsin (α1-AT) acts as a suicide inhibitor of a wide range of serine proteases. In normal humans, more than 2 g of α1-AT protein is synthesized daily, resulting in a serum concentration of about 2 mg/mL. The primary function of α1-AT protein is inhibition of neutrophil elastase, which may protect from excessive tissue damage [47]. In addition to its well established role of neutralizing elastase activity, in cell lines of both mesenchymal and epithelial origin, α1-AT can enhance DNA synthesis and cell proliferation particularly when insulin is present [46]. The highest amounts of α1-AT were clearly present in the fetal sheep liver, regardless of the stage of gestation. The decline in hepatic α1-AT in term and neonatal animals occurs at a time when the plasma levels of glucocorticoids markedly suggesting that the natural increase in cortisol during late gestation in the sheep fetus may be responsible for this decline in the abundance of hepatic alpha-1-antitrypsin. It may influence the ability of fetus to control proteinases at the fetal maternal interface and influence events leading to parturition, and/or the ultimate demise of the placenta [49].
Disruptions in pancreatic functions cause indigestion problems and altered glucose homeostasis, which can prelude the development of diabetes mellitus, pancreatic cancer and pancreatitis. The pig pancreas is an important resource of xenotransplantation for human diabetic patients, but little is known about its developmental stage at the molecular level. We presented proteomic alterations in pig pancreas by analyzing the changes in protein expression level during development. This study will provide basis for rudimentary understanding development of pig pancreas at the molecular level.
Acknowledgments
This work was supported by the grants from the Ministry of Agriculture, Food and Rural Affairs (311054-03-2-HD110) and Ministry of Science, ICT and Future Planning (2013M3A9D5072550), Korea to Prof. Je Kyung Seong.
References
1. Rao MS, Reddy JK. Pancreatic stem cells: differentiation options. Stem Cell Rev. 2005; 1(3):265–271. PMID: 17142864.

2. Portela-Gomes GM, Hacker GW, Weitgasser R. Neuroendocrine cell markers for pancreatic islets and tumors. Appl Immunohistochem Mol Morphol. 2004; 12(3):183–192. PMID: 15551729.

3. Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004; 27(5):1047–1053. PMID: 15111519.

4. Herber S, Grus FH, Sabuncuo P, Augustin AJ. Two-dimensional analysis of tear protein patterns of diabetic patients. Electrophoresis. 2001; 22(9):1838–1844. PMID: 11425240.

5. Sabek OM, Marshall DR, Penmetsa R, Scarborough O, Gaber AO. Examination of gene expression profile of functional human pancreatic islets after 2-week culture. Transplant Proc. 2006; 38(10):3678–3679. PMID: 17175365.

6. Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007; 120(5):1046–1054. PMID: 17149698.

7. Højlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J Biol Chem. 2003; 278(12):10436–10442. PMID: 12531894.
8. Qiu L, List EO, Kopchick JJ. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol Cell Proteomics. 2005; 4(9):1311–1318. PMID: 15961380.

9. Silva D, Petrovsky N. Identification of key beta cell gene signaling pathways involved in type 1 diabetes. Ann N Y Acad Sci. 2004; 1037:203–207. PMID: 15699518.
10. Ibrahim Z, Busch J, Awwad M, Wagner R, Wells K, Cooper DK. Selected physiologic compatibilities and incompatibilities between human and porcine organ systems. Xenotransplantation. 2006; 13(6):488–499. PMID: 17059572.

11. Levy MF. Animal organs for human transplantation: how close are we? Proc (Bayl Univ Med Cent). 2000; 13(1):3–6. PMID: 16389317.

12. Starzl TE, Fung J, Tzakis A, Todo S, Demetris AJ, Marino IR, Doyle H, Zeevi A, Warty V, Michaels M, Kusne S, Rudert WA, Trucco M. Baboon-to-human liver transplantation. Lancet. 1993; 341(8837):65–71. PMID: 8093402.

13. Levy MF, Crippin J, Sutton S, Netto G, McCormack J, Curiel T, Goldstein RM, Newman JT, Gonwa TA, Banchereau J, Diamond LE, Byrne G, Logan J, Klintmalm GB. Liver allotransplantation after extracorporeal hepatic support with transgenic (hCD55/hCD59) porcine livers: clinical results and lack of pig-to-human transmission of the porcine endogenous retrovirus. Transplantation. 2000; 69(2):272–280. PMID: 10670638.
14. Paradis K, Langford G, Long Z, Heneine W, Sandstrom P, Switzer WM, Chapman LE, Lockey C, Onions D, Otto E. The XEN 111 Study Group. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science. 1999; 285(5431):1236–1241. PMID: 10455044.

15. Steiner S, Anderson NL. Pharmaceutical proteomics. Ann N Y Acad Sci. 2000; 919:48–51. PMID: 11083096.

16. Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000; 21(6):1037–1053. PMID: 10786879.

17. Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1999; 34(2):105–116. PMID: 10093212.

18. Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE, Carr SA, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998; 826(2):167–181. PMID: 9871337.

19. Lainé J, Pelletier G, Grondin G, Peng M, LeBel D. Development of GP-2 and five zymogens in the fetal and young pig: biochemical and immunocytochemical evidence of an atypical zymogen granule composition in the fetus. J Histochem Cytochem. 1996; 44(5):481–499. PMID: 8627005.

20. Leblond FA, Morisset J, LeBel D. Alterations of pancreatic growth and of GP-2 content in the reserpinized rat model of cystic fibrosis. Pediatr Res. 1989; 25(5):478–481. PMID: 2717264.

21. Hayashi K, Pan Y, Shu H, Ohshima T, Kansy JW, White CL 3rd, Tamminga CA, Sobel A, Curmi PA, Mikoshiba K, Bibb JA. Phosphorylation of the tubulin-binding protein, stathmin, by Cdk5 and MAP kinases in the brain. J Neurochem. 2006; 99(1):237–250. PMID: 16925597.

22. Malorni L, Cacace G, Cuccurullo M, Pocsfalvi G, Chambery A, Farina A, Di Maro A, Parente A, Malorni A. Proteomic analysis of MCF-7 breast cancer cell line exposed to mitogenic concentration of 17beta-estradiol. Proteomics. 2006; 6(22):5973–5982. PMID: 17051647.
23. Sewell DA, Yuan CX, Robertson E. Proteomic signatures in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2007; 69(2):77–84. PMID: 17127822.

24. Zhang HZ, Wang Y, Gao P, Lin F, Liu L, Yu B, Ren JH, Zhao H, Wang R. Silencing stathmin gene expression by survivin promoter-driven siRNA vector to reverse malignant phenotype of tumor cells. Cancer Biol Ther. 2006; 5(11):1457–1461. PMID: 17012855.

25. Alli E, Yang JM, Hait WN. Silencing of stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene. 2007; 26(7):1003–1012. PMID: 16909102.

26. Ueda M, Hayase Y, Mashiba S. Establishment and evaluation of 2 monoclonal antibodies against oxidized apolipoprotein A-I (apoA-I) and its application to determine blood oxidized apoA-I levels. Clin Chim Acta. 2007; 378(1-2):105–111. PMID: 17174291.

27. Kim KD, Lim HY, Lee HG, Yoon DY, Choe YK, Choi I, Paik SG, Kim YS, Yang Y, Lim JS. Apolipoprotein A-I induces IL-10 and PGE2 production in human monocytes and inhibits dendritic cell differentiation and maturation. Biochem Biophys Res Commun. 2005; 338(2):1126–1136. PMID: 16259956.

28. Zannis VI, Cole FS, Jackson CL, Kurnit DM, Karathanasis SK. Distribution of apolipoprotein A-I, C-II, C-III, and E mRNA in fetal human tissues. Time-dependent induction of apolipoprotein E mRNA by cultures of human monocyte-macrophages. Biochemistry. 1985; 24(16):4450–4455. PMID: 3931677.

29. Abelev GI. Alpha-fetoprotein as a marker of embryo-specific differentiations in normal and tumor tissues. Transplant Rev. 1974; 20(0):3–37. PMID: 4135841.

30. Cohen BL, Orn A, Gronvik KO, Gidlund M, Wigzell H, Murgita RA. Suppression by alpha-fetoprotein of murine natural killer cell activity stimulated in vitro and in vivo by interferon and interleukin 2. Scand J Immunol. 1986; 23(2):211–223. PMID: 2419966.

31. Murgita RA, Goidl EA, Kontiainen S, Beverley PC, Wigzell H. Adult murine T cells activated in vitro by alpha-fetoprotein and naturally occurring T cells in newborn mice: identity in function and cell surface differentiation antigens. Proc Natl Acad Sci U S A. 1978; 75(6):2897–2901. PMID: 78493.

32. Alpert E, Dienstag JL, Sepersky S, Littman B, Rocklin R. Immunosuppressive characteristics of human AFP: effect on tests of cell mediated immunity and induction of human suppressor cells. Immunol Commun. 1978; 7(2):163–185. PMID: 77249.
33. Peck AB, Murgita RA, Wigzell H. Cellular and genetic restrictions in the immunoregulatory activity of alpha-fetoprotein.III. Role of the MLC-stimulating cell population in alpha-fetoprotein-induced suppression of T cell-mediated cytotoxicity. J Immunol. 1982; 128(3):1134–1140. PMID: 6173421.
34. Murgita RA, Tomasi TB Jr. Suppression of the immune response by alpha-fetoprotein on the primary and secondary antibody response. J Exp Med. 1975; 141(2):269–286. PMID: 46267.

35. Yachnin S. Demonstration of the inhibitory effect of human alpha-fetoprotein on in vitro transformation of human lymphocytes. Proc Natl Acad Sci U S A. 1976; 73(8):2857–2861. PMID: 60761.

36. Auer IO, Kress HG. Suppression of the primary cell-mediated immune response by human alpha1-fetoprotein in vitro. Cell Immunol. 1977; 30(1):173–179. PMID: 67903.
37. Littman BH, Alpert E, Rocklin RE. The effect of purified alpha-fetoprotein on in vitro assays of cell-mediated immunity. Cell Immunol. 1977; 30(1):35–42. PMID: 67905.
38. Peck AB, Murgita RA, Wigzell H. Cellular and genetic restrictions in the immunoregulatory activity of alpha-fetoprotein. I. Selective inhibition of anti-Ia-associated proliferative reactions. J Exp Med. 1978; 147(3):667–683. PMID: 75940.

39. Olinescu A, Laky M, Pofescu DE, Dumitrescu A, Ganea D. The effect of alpha-fetoprotein on the immune response. III. Diminution of the phagocytosis capacity of macrophages cultured in vitro in the presence of mouse amniotic fluid or alpha-fetoprotein. Arch Roum Pathol Exp Microbiol. 1977; 36(3-4):247–253. PMID: 81666.
40. The Shanghai coordinating group. Studies on alpha-fetoprotein v. influence of alpha-fetoprotein on immunological function. In advances in medical oncology, research, and education. In : Fox M, editor. Biological basis for cancer diagnosis. New York: Pergamon Press;1979. p. 297–301.
41. Lu CY, Changelian PS, Unanue ER. Alpha-fetoprotein inhibits macrophage expression of Ia antigens. J Immunol. 1984; 132(4):1722–1727. PMID: 6199409.
42. Crainie M, Semeluk A, Lee KC, Wegmann T. Regulation of constitutive and lymphokine-induced Ia expression by murine alpha-fetoprotein. Cell Immunol. 1989; 118(1):41–52. PMID: 2463096.
43. Morris Buus R, Boockfor FR. Transferrin expression by placental trophoblastic cells. Placenta. 2004; 25(1):45–52. PMID: 15013638.

44. Briggs DA, Sharp DJ, Miller D, Gosden RG. Transferrin in the developing ovarian follicle: evidence for de-novo expression by granulosa cells. Mol Hum Reprod. 1999; 5(12):1107–1114. PMID: 10587364.

45. Lampreave F, Pineiro A. Concentrations of major plasma proteins in serum and whole-tissue extracts of porcine fetuses during development. J Reprod Fertil. 1992; 95(2):441–449. PMID: 1381439.

46. Nahreini P, Hovland AR, Kumar B, Andreatta C, Edwards-Prasad J, Prasad KN. Effects of altered cyclophilin A expression on growth and differentiation of human and mouse neuronal cells. Cell Mol Neurobiol. 2001; 21(1):65–79. PMID: 11440199.
47. Jang G, Bhuiyan MM, Jeon HY, Ko KH, Park HJ, Kim MK, Kim JJ, Kang SK, Lee BC, Hwang WS. An approach for producing transgenic cloned cows by nuclear transfer of cells transfected with human alpha 1-antitrypsin gene. Theriogenology. 2006; 65(9):1800–1812. PMID: 16303172.

48. She QB, Mukherjee JJ, Crilly KS, Kiss Z. alpha(1)-antitrypsin can increase insulin-induced mitogenesis in various fibroblast and epithelial cell lines. FEBS Lett. 2000; 473(1):33–36. PMID: 10802054.
49. Berdusco ET, Yang K, Challis JR, Hammond GL. Tissue distribution of alpha 1-proteinase inhibitor messenger ribonucleic acid and its regulation by glucocorticoids in fetal and neonatal sheep. Biol Reprod. 1993; 49(4):816–821. PMID: 8218647.
Figure 1
Representative Coomassie blue stained 2-DE gels of pig pancreas based on developmental stages. The proteins (1 mg) were separated on a 24 cm, pH 3-10 nonlinear strip, followed by an 8-18% gradient SDS-PAGE. Proteins were detected by Coomassie blue staining on the 2-DE images of the master gel. The proteins identified are designated with their accession numbers. (A) E60, (B) neonate, (C) adult pig pancreas. The symbol at (C) indicates development group number (1, 2 and 3) and alphabet order in Table 1. Mr indicates molecular weight of proteins.
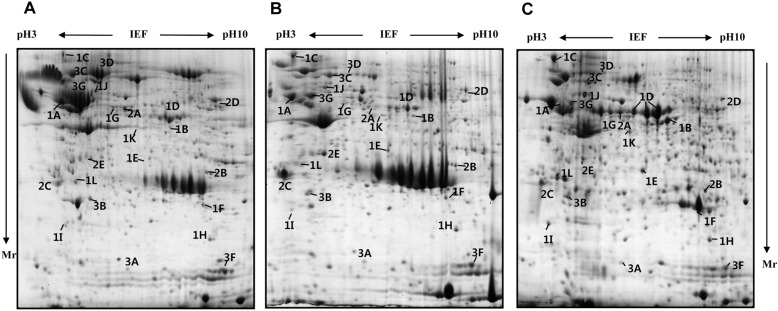
Figure 2
2-DE gel enlargements showing proteins that are highly expressed in the adult stage. Various zymogens (pancreatic alpha-amylase precursor, trypsin precursor and pancreatic triacylglycerol lipase) and protein synthesis related proteins (protein disulfide-isomerase precursor, endoplasmin precursor, 60 kDa heat shock protein and phosphatidylethanolamine-binding protein) are up-regulated. The proteins of interest are indicated by arrows. These proteins were identified by MALDI-TOF MS (A. protein disulfide-isomerase precursor, B. glycine amidinotransferase, C. endoplasmin precursor, D. pancreatic alpha-amylase precursor, E. MHC class II antigen, F. trypsin precursor, G. pancreatic triacylglycerol lipase, H. phosphatidylethanolamine-binding protein, I. translationally-controlled tumor protein, J. 60 kDa heat shock protein, K. hypothetical protein FLJ39175, and L. laminin receptor).
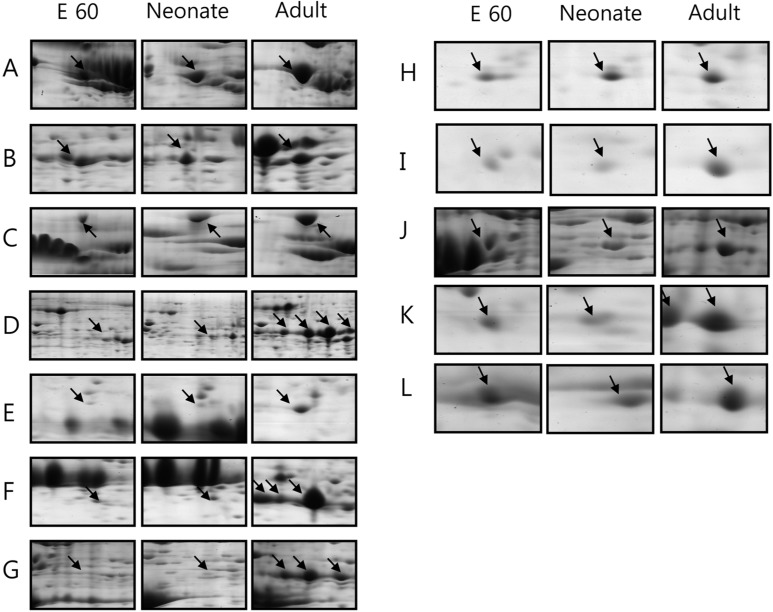
Figure 3
2-DE gel enlargements showing the proteins especially expressed in the neonatal stage. Chymotrypsinogen 2 and 14-3-3 protein are significantly up-regulated at birth. The proteins of interest are indicated by arrows. These proteins were identified by MALDI-TOF MS (A. keratin, 53K type II cytoskeletal, B. chymotrypsinogen 2, C. 14-3-3 protein, D. pyruvate kinase (EC 2.7.1.40) isozyme M2, E. spermidine synthase (EC 2.5.1.16)).
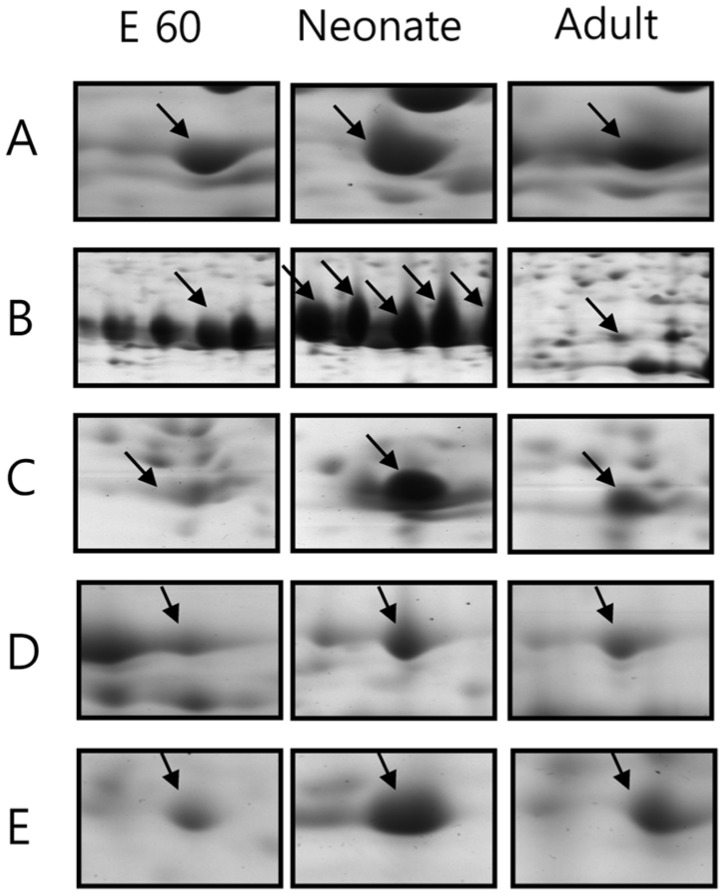
Figure 4
2-DE gel enlargements showing the proteins especially more expressed in the embryonic stage. Apolipoprotein A-I precursor, alpha-fetoprotein, transferrin and alpha-1-antitrypsin precursor are markedly expressed in fetal developing pancreas. The proteins of interest are indicated by arrows. These proteins were identified by MALDI-TOF MS (A. stathmin (Phosphoprotein p19), B. apolipoprotein A-I precursor, C. alpha-fetoprotein, D. transferrin. E. F25B3.3 kinase like protein (Fragment), F. peptidyl-prolyl cis-trans isomerase A, G. alpha-1-antitrypsin precursor).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download