Abstract
Loss-of function mutations in the transmembrane inner ear expressed (Tmie/TMIE) gene have been shown to cause deafness in mice and humans (DFNB6). Previous studies report that the circling mouse can be an animal model for DFNB6. However, the expression pattern of Tmie protein in postnatal developmental stages has not been clearly revealed. In this study we tried to investigate the expression of Tmie protein in the liver, spleen, kidney, and lung, as well as in the cochlea. We examined various tissue samples from five different age groups of C57BL/6J animals. Using western blotting analysis, the expression of Tmie protein in these organs has been identified. The results show that Tmie protein expression in the cochlea has been increased in postnatal developmental stages, indicating that Tmie plays an important role in not only the development and also in the function of the cochlea. The expression pattern of Tmie in adult mouse organs such as the liver, spleen, kidney, and spleen significantly vary in adult rats. The order of Tmie expression level in mice (63 days after birth) was spleen, liver, lung, cochlea, and kidney, whereas in the adult rat it was liver, cochlea, lung, spleen, and kidney.
The incidence of hearing loss in humans is substantial, with a frequency of pre-lingual deafness as high as 0.1-0.2% and a similar frequency of post-lingual deafness before the third decade of life [1,2]. In developed nations, ~50% of these cases appear to have a genetic basis. Inherited deafness in humans is genetically heterogeneous, with effects in any one of more than 100 distinct genes likely to be responsible for nonsyndromic hearing loss [1]. Despite the difficulty inherent in the analysis of genetically heterogeneous conditions, there has been dramatic progress in the localization and identification of a large number of genes associated with hearing loss during the past several years [3-5]. Mutations in 15 different genes, including the transmembrane inner ear (TMIE), have been shown to cause nonsyndromic, recessively inherited hearing loss in humans [3]. Recently, loss of function mutations in the Tmie/TMIE gene has been shown to cause deafness in humans (DFNB6). These results indicate that the Tmie/TMIE gene has a conserved, critical role in the auditory system [6,7]. Mouse genetic models provide a valuable approach to identify genes which play a role in inherited human hearing loss, and also offer a useful system to investigate gene function [8]. Identification and analysis of these genes in mice has implicated a diverse array of proteins required in the inner ear during early embryonic development and postnatal maturation of the sensory neuroepithelium, as well as in the adult organ [9]. The Tmie gene has no sequence similarity with other known genes and their functions are presently unclear. Previously our research group reported that the circling mouse is a possible animal model for deafness and it has a 40 kb deletion that includes the Tmie [10,11]. On the basis of these results, circling mice are believed to be an excellent animal model for investigating inner ear abnormalities in humans. However the functional roles of Tmie in the cochlea and other organs such as the liver, lung, kidney and spleen remain unclear. The levels of expression, the distribution and the time course of Tmie have not been defined. Only the cochlear pathology of circling mice and the expression of Tmie protein in various organs of adult rats have been analyzed [10,12]. All these results led us to investigate the expression pattern of Tmie protein in the postnatal developmental stages of C57BL/6J mice.
C57BL/6J mice were used throughout this study. C57BL/6J mice were obtained from the Korea Research Institute of Bioscience and Biotechnology (Deajon, South Korea) and bred in our facilities. The mice were kept in a specific-pathogen free animal care facility and were housed individually in plastic cages (18×30×15 cm) with corn cob bedding. The facility was maintained at 22±2℃ temperatures, in 55±10% relative humidity, and a 12 hr light and 12 hr dark routine cycle was employed. Normal rodent pellet diet (Jeiljedang, Seoul, Korea) and water was supplied ad libitum. The animal study was conducted in accordance with the guidelines and with the approval of the Institutional Animal Care and Use Committee of Hallym University (Hallym-1-51).
Tissue samples (kidney, liver, lung, spleen, and cochlea) were harvested from mice of different age groups (7, 21.42, and 63). We were used western blotting analysis for the expression of Tmie protein in various organs in different age groups. The tissue homogenates (whole cell extract) were prepared by the sonication of these tissues in lysis buffer (50 mM Tris-HCl, pH 6.8, 100 mM DTT, 2% SDS, 10% glycerol). These tissue homogenates (50 µg of the whole cell extract) were boiled for 5 min and then resolved by SDS PAGE electrophoresis (15% gel) runs at 200 V for 60 min in a running buffer (25 mM Tris, 192 mM Glycin, 0.1% SDS). Electrophoretic transfer to a polyvinylidene fluoride transfer membrane (PVDF, Pall Corporation, USA) was then carried out at 100 V for 90 min using the transfer buffer according to the manufacturer's instructions. The membrane was blocked with 5% BSA in PBS at room temperature for 1 h, then incubated for overnight at 4℃ with the anti-Tmie antibody diluted to 1/1,000 in 1% BSA buffer, followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Molecular Probes, USA) for an additional 1 h. The blot was finally developed using Luminol (PerkinElmer Life Sciences, Inc., USA) and then examined with FUJIFILM Luminescent Image Analyzer and Image Reader LAS-1000 Lite software (Fuji Photo Film Co., Ltd, Japan).
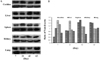
Tmie expression in various mouse organs, including the cochlea, liver, spleen, kidney, and lung, has been identified by using the anti-Tmie antibody which was previously developed by our group [12,13]. A single immunoreactive band for Tmie protein was found in the cochlea, liver, spleen, kidney, and lung tissue samples (Figure 1). The blot was also identified with an anti-β actin antibody to determine the relative level of protein in each lane. The relative expression level of Tmie protein in each organ was determined by the ratio of the intensity of the immunoreactive bands against the anti-Tmie and anti-β actin antibodies. As shown in Figure 1A, the expression level of Tmie protein in the cochlea increased throughout the postnatal developmental stages, indicating that Tmie plays an important role in the development and function of the cochlea. In addition, there was the highest expression level of Tmie protein at 42 days of age in the liver. The expression level of Tmie protein in the spleen and kidney slightly decreased throughout the postnatal developmental stages. In the lung, Tmie protein expression increased throughout the study period. Figure 1B show ratios of the intensities of the immunoreactive bands against anti-Tmie and anti-β actin antibodies. The order of Tmie expression level was cochlea, kidney, liver, lung, and spleen at 7 days after birth (cochlea 1.6, spleen 0.4). This pattern of Tmie expression level changed with later stages of mice. At 63 days after birth, the order of Tmie expression level was spleen, liver, lung, cochlea, and kidney. The Tmie expression level in several organs of adult rats has been reported previously [15]. Order of Tmie expression level in adult rats was liver, cochlea, lung, spleen, and kidney.
To know features of Tmie protein, we analyzed Tmie protein using UniProt analysis (Figure 2). Tmie protein has a signal peptide (1-28 AA), extracellular region (29-58 AA), transmembrane region (59-79 AA), cytoplasmic region (80-153 AA) and lycine-rich domain (124-153 AA). Tmie protein is need for correct development of stereocilia bundles in the cochlea [16]. The mature Tmie protein is localized in the plasma membrane [13]. From features of Tmie protein by UniProt analysis and previous results, Tmie protein may reside within an internal membrane compartment and function in vesicle trafficking.
There have been a few reports that deafness genes have been involved with cancer and liver transplantation patients having hearing impairment side effects [17,18]. As a primary step to reveal the deafness proteins related with other organs, we examined the level of Tmie protein expression in these organs. Taken together, these results show that Tmie protein could be important and that it could play a role in the function of these organs on the basis of the needs of each particular organ. More functional studies including localization of Tmie protein in liver, spleen, lung, and kidney should are needed to understand the function of Tmie in these organs.
Figures and Tables
Figure 1
Western blot analysis of Tmie expression in C57BL/6J mice. A) Tmie expression has been identified by using the anti-Tmie antibody which was previously developed by our group [12]. A single immunoreactive band of approximately 26 kDa was observed in the cochlea, liver, spleen, kidney, and lung tissue (upper panel of each tissue). The amount of protein loaded for each lane was assessed by determining the presence of 43 kDa β-actin (lower panel of each tissue). B) Intensities from the immunoreactive bands against anti-Tmie and anti-â actin antibodies were calculated and then plotted to show the relative levels of Tmie in postnatal developmental stages. Results of two independent experiments are shown.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (KRF-2007-521-E00140), and by Priority Research Centers Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2011-0030750).
References
1. Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991. 630:16–31.
2. Mehl AL, Thomson V. Newborn hearing screening: the great omission. Pediatrics. 1998. 101(1):E4.
3. Petit C, Levilliers J, Hardelin JP. Molecular genetics of hearing loss. Annu Rev Genet. 2001. 35:589–646.
4. Resendes BL, Williamson RE, Morton CC. At the speed of sound: gene discovery in the auditory system. Am J Hum Genet. 2001. 69(5):923–935.
5. Keats BJ, Berlin CI. Genomics and hearing impairment. Genome Res. 1999. 9(1):7–16.
6. Mitchem KL, Hibbard E, Beyer LA, Bosom K, Dootz GA, Dolan DF, Johnson KR, Raphael Y, Kohrman DC. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet. 2002. 11(16):1887–1898.
7. Chung WH, Kim KR, Cho YS, Cho DY, Woo JH, Ryoo ZY, Cho KI, Hong SH. Cochlear pathology of the circling mouse: a new mouse model of DFNB6. Acta Otolaryngol. 2007. 127(3):244–251.
8. Probst FJ, Camper SA. The role of mouse mutants in the identification of human hereditary hearing loss genes. Hear Res. 1999. 130(1-2):1–6.
9. Steel KP, Kros CJ. A genetic approach to understanding auditory function. Nat Genet. 2001. 27(2):143–149.
10. Lee JW, Ryoo ZY, Lee EJ, Hong SH, Chung WH, Lee HT, Chung KS, Kim TY, Oh YS, Suh JG. Circling mouse, a spontaneous mutant in the inner ear. Exp Anim. 2002. 51(2):167–171.
11. Cho KI, Suh JG, Lee JW, Hong SH, Kang TC, Oh YS, Ryoo ZY. The circling mouse (C57BL/6J-cir) has a 40-kilobase genomic deletion that includes the transmembrane inner ear (tmie) gene. Comp Med. 2006. 56(6):476–481.
12. Nam YY, Karuppasamy S, Park BK, Kwon HJ, Suh JG. Production and characterization of polyclonal antibody to transmembrane inner ear protein. Lab Anim Res. 2009. 25(2):145–150.
13. Karuppasamy S, Nam YY, Jung H, Park B, Kwon HJ, Suh JG. Subcellular localization of the transmembrane inner ear (Tmie) protein in a stable Tmie-expressing cell line. Lab Anim Res. 2011. 27(4):339–342.
14. Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, Ramesh A, Srisailpathy S, Deshmukh D, Riazuddin S, Griffith AJ, Friedman TB, Smith RJ, Wilcox ER. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet. 2002. 71(3):632–636.
15. Su MC, Yang JJ, Chou MY, Hsin CH, Su CC, Li SY. Expression and localization of Tmie in adult rat cochlea. Histochem Cell Biol. 2008. 130(1):119–126.
16. Chung WH, Kim KR, Cho YS, Cho DY, Woo JH, Ryoo ZY, Cho KI, Hong SH. Cochlear pathology of the circling mouse: a new mouse model of DFNB6. Acta Otolaryngol. 2007. 127(3):244–251.
17. Op de Beeck K, Van Camp G, Thys S, Cools N, Callebaut I, Vrijens K, Van Nassauw L, Van Tendeloo VF, Timmermans JP, Van Laer L. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur J Hum Genet. 2011. 19(9):965–973.
18. Rifai K, Kirchner GI, Bahr MJ, Cantz T, Rosenau J, Nashan B, Klempnauer JL, Manns MP, Strassburg CP. A new side effect of immunosuppression: high incidence of hearing impairment after liver transplantation. Liver Transpl. 2006. 12(3):411–415.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download