Abstract
The drug resistance of microorganisms isolated from laboratory animals never treated with antibiotics is being reported consistently, while the number of laboratory animals used in medicine, pharmacy, veterinary medicine, agriculture, nutrition, and environmental and health science has increased rapidly in Korea. Therefore, this study examined the development of antimicrobial resistance in bacteria isolated from laboratory animals bred in Korea. A total of 443 isolates (7 species) containing 5 Sphingomonas paucimobilis, 206 Escherichia coli, 60 Staphylococcus aureus, 15 Staphylococcus epidermidis, 77 Enterococcus faecalis, 27 Citrobacter freundii, 35 Acinetobacter baumannii were collected from the nose, intestine, bronchus and reproductive organs of ICR mice and SD rats. Of these species, Acinetobacter baumannii and Enterococcus faecalis showed significant antimicrobial resistance according to the minimum inhibition concentration (MIC) in E-test. In case of Acinetobacter baumannii, several isolates showed MIC values 16-128 µg/mL for cefazolin and cefoxitin, and higher resistance (128-512 µg/mL) to nitrofurantoin than that of standard type. Resistance to cefazolin, cefoxitin and nitrofurantoin was detected in 17.14, 20.00, and 8.57% of the Acinetobacter baumannii isolates, respectively. In addition, 44.1% of the Enterococcus faecalis isolates collected from the laboratory animals were resistant to oxacillin concentration of 16-32 µg/mL range, while MIC value of standard type was below oxacillin concentration of 6 µg/mL. These results suggest that in rodent species of laboratory animals, Acinetobacter baumannii are resistance to cefazolin, cefoxitin and nitrofurantoin, whereas those of Enterococcus faecalis were resistance to oxacillin.
Various antibiotics have been administered to animals in order to accomplish a range of purposes including growth promotion, prophylaxis and therapy. These administrations lead to the appearance and accumulation of novel resistance bacteria in their flora [1]. In particular, the large number of drug-resistance organisms found in farm animals is a major cause of resistance in human and other animals [2]. Of the various type animals, laboratory animals showed a low incidence of drug resistance because antibiotics have not been used for the breeding of laboratory animals for a long time [3]. Furthermore, the bacterial isolates from laboratory animals showed a different pattern of drug resistance from those in the human population [4]. Therefore, it is important to isolate and characterize antibiotic-resistant bacteria from laboratory animals to prevent microbial contamination of the facility and improve the efficiency of post-operative care in a laboratory. This study monitored the development of antibiotics resistance in bacteria isolated from laboratory animals, which had never been treated with antibiotics.
The animal protocol used in this study was reviewed and approved based on the ethical procedures and scientific care by the Korea FDA-Institutional Animal Care and Use Committee (PNU-IACUC; Approval Number 1101KFDA07). Adult ICR mice and SD rats were handled in an accredited Korea Food & Drug Administration (Korea FDA) animal facility in accordance with the AAALAC International Animal Care policies (Accredited Unit-Korea Food and Drug Administration: Unit Number-000996). All animals were given a standard irradiated chow diet (Purina Mills, Seoungnam, Korea) ad libitum, and maintained in a specific pathogen-free state under a strict light cycle (light on at 06:00 h and off at 18:00 h) at 22±2℃ and 40-60% relative humidity.
A total of 443 isolates of 7 species microorganisms were collected from representative infection organs, such as the nose, intestine, bronchus and reproductive organs, of 24 laboratory animals (12 ICR mice and 12 SD rats) bred in Korea FDA facility using streak plate methods. After isolation of each colony on conventional bacteria media, isolates were identified with Vitek2 compact autoanalyzer system manufactured by BioMerieux (NC, USA). All identification procedures were performed according to manufacturer's manual and applicated Vitek2-GPI and -GNI cards for testing. From identification results, ID results with a single acceptable identification were accepted as suggested ID results. Furthermore, the species of isolates identified by Vitek2 system were conformed with the biochemical test using API kit (BioMerieux).
As listed Table 1, a total of 443 isolates were spread differentially throughout 7 different bacteria species. The most prevalent microorganisms were Escherichia coli (46.6%), Enterococcus faecalis (17.3%), Staphylococcus aureus (13.6%) and Acinetobacter baumannii (8.0%) followed by Staphylococcus epidermidis (7.4%), Citrobacter freundii (6.0%) and Sphingomonas paucimobilis (1.1%). A larger number of isolates was found in ICR mice than SD rats, even though the total number of Staphylococcus epidermidis isolates in rats was higher than that in mice.
The antimicrobial resistances were determined in all isolates using the minimum inhibition concentration (MIC) method [5]. All isolates were E-tested for susceptibility to amikacin, amoxicillin/clavulanic acid, ampicillin, cefazolin, cefepime, cefotaxime, cefoxitin, cefbazidime, ciprofloxacin, imipenem, netilmicin, nitrofurantoin, nortfloxaci, piperacillin, piperacillin/tazobactam, tetracycline and oxacillin by E-strip.
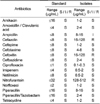
Of the 7 species, 2 species (Acinetobacter baumannii and Enterococcus faecalis) that showed a high incidence of drug resistance in laboratory animals were selected. Acinetobacter baumannii belongs to Acinetobacter genus, and is an opportunistic pathogen responsible for nosocomial infections and is associated with an elevated human mortality rate [6]. In addition, most isolates of Acinetobacter baumannii showed multidrug resistance because they contained a small genome, islands of alien DNA, and other cytological and genetic materials [7]. In this study, the drug resistance of Acinetobacter baumannii was detected in only three antibiotics, cefazolin, cefoxitin and nitrofurantoin, among the 16 antibiotics used in the test. Nitrofurantoin was the most efficient (128-512 µg/mL), whereas the same percentages of resistance (16-128 µg/mL) were found against cefazolin and cefoxitin. However, Acinetobacter baumannii was not showed any specific resistance to other antibiotics (Table 2). Furthermore, nitrofurantoin and cefazolin resistance appear to be uniform with only slight differences between the isolates from ICR mice and SD rats. Cefoxitin resistance, however, was more prevalent in the isolates from SD rats than in those from ICR mice. In case of the nitrofurantoin resistance, the resistance carrier of isolates was never detected in the male SD rats (Table 3). These results are very different from those of animals slaughtered for human consumption. Sixteen isolates of Acinetobacter baumannii isolated from livestock were sensitive to carbapenems, gentamicin, ciprofloxacin and piperacillin/tazobactam but were resistant to amoxicillin, cefradine, trimethoprim and chloramphenicol [8]. On the other hand, the isolates of Acinetobacter baumannii isolated from laboratory animals were sensitive only to cefazolin, cefoxitin and nitrofurantoin. Therefore, these findings showed that Acinetobacter baumannii isolated from laboratory animals were a bacterium which were resistant to treatment by the novel range of antibiotics.
Oxacillin resistance was also detected in Enterococcus faecalis among 7 species isolated from laboratory animals. Enterococcus faecalis could cause life-threatening infections in the nosocomial environment of humans, where the naturally high levels of antibiotic resistance found in Enterococcus faecalis contribute to its pathogenicity [9]. Enterococcus faecalis is resistant to many commonly used antimicrobial agents (aminoglycosides, aztreonam, cephalosporins, clindamycin, semisynthetic penicillins [nafcillin and oxacillin], and trimethoprim-sulfamethoxazole). Resistance to vancomycin in Enterococcus faecalis is becoming increasingly widespread [10,11]. The treatment options for vancomycin-resistant Enterococcus faecalis include linezolid and daptomycin; ampicillin is preferred if the bacteria are susceptible [12]. In this study, the isolates of Enterococcus faecalis showed a higher resistance against oxacillin than that of standard type (Figure 1A), while Staphylococcus aureus and Staphylococcus epidermidis were not resisted to treatment by the usual range of oxacillin (Figure 1B). In addition, the inhibition concentration of Enterococcus faecalis isolates was the 6-32 µg/mL of oxacillin, but the inhibition concentration of standard type was below 6 µg/mL of oxacillin (Table 4). Furthermore, oxacillin resistance was observed frequently in Enterococcus faecalis isolated from mice with 25 (52.00%) isolates of the 48 Enterococcus faecalis carriers than those of rats (31.00%). The isolates in female ICR mice (65.00%) and SD rats (35.20%) showed high levels of resistance compared to those (42.80% and 25.00%) of male animals (Table 5). These results are in agreement with previous reports and provide novel evidence of high levels of oxacillin resistance in Enterococcus faecalis from ICR mice and SD rats bred in Korea.
In conclusion, these results firstly showed an evidence that the drug resistance of microorganisms collected from laboratory animals had occurred in breeding facility of Korea. Especially, Acinetobacter baumannii and Enterococcus faecalis isolated from ICR mice and SD rats were representative microorganisms having resistance against antibiotics.
Figures and Tables
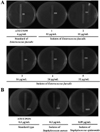 | Figure 1MIC test of Enterococcus faecalis resistance to oxacillin. The drug resistance isolates were sensitive in range from 16 to 32 µg/mL of oxacillin, even though the standard microorganism (ATCC 5199) was quite sensitive to more than 6 µg/mL of oxacillin. (A) Oxacillin resistance of Enterococcus faecalis isolates collected from ICR mice or SD rats. (B) Oxacillin sensitivity of Staphylococcus aureus and Staphylococcus epidermidis isolates collected from ICR mice or SD rats. |
Table 2
Resistance pattern of the Acinetobacter baumannii isolates collected from laboratory mice and rats never treated with antibiotics
References
1. DuPont HL, Steele JH. Use of antimicrobial agents in animal feeds: implications for human health. Rev Infect Dis. 1987. 9(3):447–460.
2. Kruse H, Sorum H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl Environ Microbiol. 1994. 60(11):4015–4021.
3. Maejima K, Urano T, Tamura H, Terakado N. Drug resistance of organisms isolated from feces of laboratory mice and rats. Jikken Dobutsu. 1980. 29(1):71–75.
4. Hansen AK, Velschow S. Antibiotic resistance in bacterial isolates from laboratory animal colonies naive to antibiotic treatment. Lab Anim. 2000. 34(4):413–422.
5. Baker CN, Banerjee SN, Tenover FC. Evaluation of Alamar colorimetric MIC method for antimicrobial susceptibility testing of gram-negative bacteria. J Clin Microbiol. 1994. 32(5):1261–1267.
6. Song JY, Cheong HJ, Choi WS, Heo JY, Noh JY, Kim WJ. Clinical and microbiological characterization of carbapenem-resistant Acinetobacter baumannii bloodstream infections. J Med Microbiol. 2011. 60(5):605–611.
7. Sullivan DR, Shields J, Netzer G. Fatal case of multi-drug resistant Acinetobacter baumannii necrotizing fasciitis. Am Surg. 2010. 76(6):651–653.
8. Hamouda A, Findlay J, Al Hassan L, Amyes SG. Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents. 2011. 38(4):314–318.
9. Rocas IN, Siqueira JF Jr, Santos KR. Association of Enterococcus faecalis with different forms of periradicular diseases. J Endod. 2004. 30(5):315–320.
10. Amyes SG. Enterococci and streptococci. Int J Antimicrob Agents. 2007. 29:Suppl 3. S43–S52.
11. Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006. 42:Suppl 1. S25–S34.
12. Arias CA, Contreras GA, Murray BE. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect. 2010. 16(6):555–562.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download