Abstract
The faded mouse is a coat color mutant that shows faded coat color and age-related loss of pigmentation. This mutation is transmitted by an autosomal recessive gene with 100% penetrance. In the present study, we carried out linkage analysis of the faded (fe) gene using intra-specific backcross panels. Affected faded mice were carefully confirmed by their faded coat color at about 4 weeks of age. In the intra-specific backcross between faded and CBA mice (n=198), the fe gene was mapped to a region 2.1 cM distal to D10mit191. Therefore, the gene order was defined as follows: centromere-D10mit51 (12.4±2.4 cM)-D10mit191 (2.1±1.0 cM)-fe-D10mit44 (13.3±2.4 cM)-D10mit42 (14.4±2.5 cM). This linkage map of the fe locus will provide a good entry point to isolate the fe gene. Since the faded mouse has pigmentary abnormalities, this mutant may be a useful model for studies of pigmentary abnormalities in humans.
The choice of the mouse as an experimental pigmentation model relates to its high level of coat color mutations (Tobin and Paus, 2001). These animals were readily amenable to genetic studies because of their observable phenotypes (Marks et al, 1999). Nearly 100 genetic loci in the mouse are known to affect coat color (Silvers, 1979). A number of the genes in which these mutations occur have now been cloned, and have led to the finding that these mutations often correspond to human genetic disease loci (Marks et al, 1999). Moreover, these mutations have increased our knowledge regarding the cellular and molecular processes involved in pigmentary abnormalities in humans. For example, dominant spotting (W) (Kunisada et al, 1998), microphthalmia (mi) (O'Reilly et al, 2001), piebald (s) (Lamoreux, 1999) and lethal spotting (ls) (Gariepy et al, 1998) mutants are animal models with mutations that affect the migration of neural crest cells. Mutations at these loci are responsible for piebaldism (Richards et al, 2001), Waardenburg syndrome 2 (Wang et al, 1998), Hirschsprung disease (Shanske et al, 2001) and Shah-Waardenburg disease (Edery et al, 1996) respectively in humans. Albino (c), W, Sl, mi, s, and ls (Yokoyama et al, 1990) provide animal models with mutations that affect melanoblast proliferation and differentiation. Mutations at the albino (c) locus are responsible for Oculocutaneous albinism I (Tomita et al, 2000). Brown (b) (Johnson and Jackson, 1992), pink-eyed dilution (p) (Fukuta et al, 1991) and recessive yellow (e) (Wada et al, 1999) provide animal models with mutations that affect the nature of the pigment produced within melanocytes. Mutations at these loci are responsible for Oculocutaneous albinism III (Toyofuku et al, 2001), Oculocutaneous albinism II (Brilliant, 2001) and red hair (Voisey et al, 2001), respectively, in humans. Grisceli disease has been shown to encode Myosin-V, which is mutated in the dilute mouse mutation. Mutations at this locus provide animal models with mutations that affect the distribution of pigment in melanocytes (Pastural et al, 1997). Genetic studies on mouse coat color mutations may yield valuable information for studies of pigmentary abnormalities in humans.
The complex melanin biosynthetic pathway is rate limited by the enzyme tyrosinase. Tyrosinase catalyzes the initial step in the melanin biosynthetic pathway: tyrosine to DOPA to DOPAquinone (Lehman et al, 2000). Tyrosinase is required for the production of both eumelanin and pheomelanin (Kobayashi et al, 1995). Hair pigmentation results from melanin synthesis and transferral by follicular melanocytes to precortical hair matrix keratinocytes, and is tightly coupled to the hair growth cycle (Slominski et al, 1994). The hair growth cycle is characterized by three developmental stages: resting (telogen), growth (anagen), and regression (catagen) (Slominski and Paus, 1993). The melanogenic activity of follicular melanocytes is strictly coupled to the anagen stage of the hair cycle. In the catagen phase, melanin formation is switched off (a decline in the activities of tyrosinase, TRP-1 and TRP-2) and is absent throughout the telogen phase (Slominski et al, 1994; Tobin and Paus, 2001).
Faded (fe) mouse is coat color mutant that was first described by (Oh et al, 1986). The mutation first occurred as a spontaneous mutation in the KSB inbred strain at Nagoya University. An autosomal recessive gene transmits this mutation with 100% penetrance. Loss of pigment occurs progressively with age, so that homozygotes are almost white by 1 year of age (Oh et al, 1986).
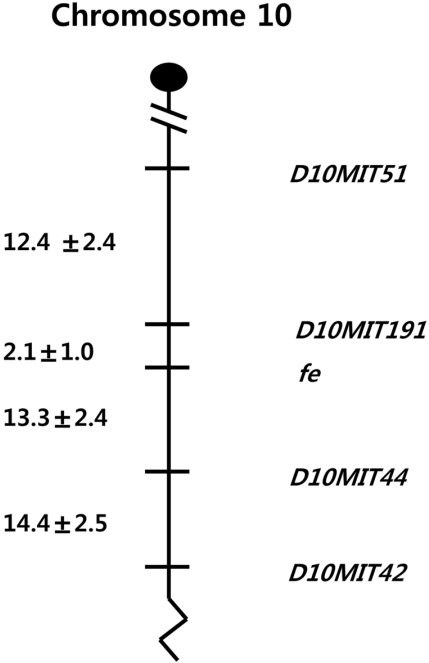
For a first step in isolating the fe gene, we report here a genetic map of fe using a backcross panel between C57BL/6J-fe/fe and CBA/J constructed by application of linkage analysis. The gene order was defined as follows: centromere-D10mit51-D10mit191-fe-D10mit44-D10mit42. This linkage map of the fe locus will provide a good entry point to isolate the fe gene.
Faded mice were maintained as a congenic strain of C57BL/6J mice (C57BL/6J-fe/fe) in the experimental animal center at Hallym University, Korea. The inbred strain CBA/J was introduced from Nagoya University, Japan, and was subsequently maintained in our laboratory. The animals were maintained at 22±2℃ temperature, in 55±10% relative humidity, and under 12-h artificial lighting from 08:30 to 20:30. Mice were given commercial laboratory chow made for mice (Jeiljedang, Seoul, Korea) and water ad libitum. This animal study was conducted in accordance with guidelines and approval of the Institutional Animal Care and Use Committees of Hallym University.
Among several kinds of hair, awls were examined because they clearly showed abundant pigment granules. Awls were gently plucked from the backs of animals and the hairs were dehydrated in alcohol and cleared in xylene. The pigment of the hair was observed under an Axioscope microscope (Carl Zeiss, Germany). Affected faded mice were carefully confirmed by their faded coat color at about 4 weeks of age.
Faded mice were crossed with the CBA/J mouse strain to produce an intra-specific cross. F1 mice were then backcrossed to faded mice, and the progeny were typed. One hundred and ninety-six intra-specific progeny were analyzed. Linkage analyses were performed using microsatellite markers by a modification of a previously described method (Suh et al, 1995). In total, 114 MIT markers, including 6 markers on each chromosome, were used for the linkage test. Genomic DNA was prepared from the tail of N2 progeny by standard SDS/proteinase K lysis and the phenol/chloroform extraction method.
Polymerase-chain reactions (PCR) using 400 ng of template DNA were carried out in a total volume of 25 µL containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 2 pmol of each primer, and 1.75 U Taq polymerase (Takara Bio Inc., Otsu, Japan). Target DNA was amplified in a Cyclogene Dri-Block® cycler (Techne Inc., Burlington, USA), with denaturation at 94℃ for 30 sec, annealing at 57-62℃ for 1 min, extension at 72℃ for 2 min, followed by a final extension at 72℃ for 7 min at the end of 30 cycles. Modifications in annealing temperature and/or magnesium concentration were made as needed to optimize the results with some primer pairs (Table 1). The amplification products were separated on 15% polyacrylamide gel and detected by ethidium bromide staining.
Percentage recombination and standard error were calculated according to Green (1981). Genetic distances are represented as cM±standard error, and order was established by minimizing the putative double crossover events.
A mapping study was performed with 196 offspring from the intra-specific backcross [(C57BL/6J-fe/fe×CBA/J)F1×C57BL/6J-fe/fe]. By typing of the 114-microsatellite markers in the N2 progeny, significant linkage was observed between the fe locus and D10mit51, 191, 44, and 42 on mouse Chromosome 10. No significant linkage was observed between the fe locus and the other microsatellite markers. Affected faded mice from the N2 progeny can be distinguished at 15 days of age by a slight dilution of coat color but are more easily classified at 30 days. Affected faded mice were carefully confirmed by their faded coat color at about 4 weeks of age (Figure 1). Loss of pigment occurs progressively with age so that homozygotes are almost white by 1 year of age.
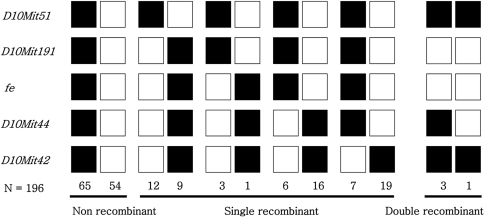
Twenty-five mice showed a recombination between fe and D10mit51 in the proximal region of the fe locus, yielding a genetic distance of 13.2±2.4 cM between these two loci. Four mice exhibited a haplotype where the recombination occurred between fe and D10mit191, indicating that D10mit191 is 2.1±1.0 cM proximal to fe. One double recombination event between fe and D10mit51 was found in the 196 N2 animals (Figure 2).
In the distal region of the fe locus, 48 mice showed a recombination between fe and D10mit42. The estimated distance between fe and D10mit42 is 26.0±3.1 cM. Twenty-two mice showed recombination between fe and D10mit44, indicating that the distance between fe and D10mit44 is 13.3±2.4 cM. Three double recombination events between fe and D10mit42 were found in the 196 N2 animals (Figure 2). Since D10mit51 has been shown to be proximal to D10mit42, the gene order was defined as follows: centromere-D10mit51 (12.4±2.4 cM)-D10mit191 (2.1±1.0 cM)-fe-D10mit44 (13.3±2.4 cM)-D10mit4 (14.4±2.5 cM) (Figure 3).
Genes that regulate pigmentation in the mouse have proved particularly useful for characterizing mutations and the way of mutations affect phenotype (Johnson and Jackson, 1992). A number of the genes in which these mutations occur have now been cloned, leading to the finding that these mutations often correspond to human genetic disease loci (Marks et al, 1999). Because the faded mice have a faded coat color and age-related loss of pigmentation, this mutation could also be used as an important model for human hereditary diseases such as various pigmentary abnormalities.
However, the gene product and function were not evident based on previous studies, and because of this, we employed linkage analysis to define the region containing the fe locus. We have thus established a linkage map in the vicinity of the fe locus by generating the intra-specific backcross panels penetrant for the faded mutation. The systematic characterization of our backcrosses provided a panel of DNAs for independent mapping of markers and gene loci. One hundred ninety-six mice [(C57BL/6J-fe/fe×CBA/J)F1×C57BL/6J-fe/fe] progenies were used for this analysis. From the CBA/J backcross, we refined our linkage map through resolution of internal SSLPs and determined the order: D10mit51, D10mit191, D10mit44 and D10mit42. The minimal genetic interval for the localization of the fe locus in this panel has been established between the markers D10mit191 and D10mit44, covering 14.2±2.4 cM.
The fe gene is linked to Ggc and Ldr, which are located on mouse chromosome 6 (Oh and Tomita, 1987). However, this knowledge was not incorporated into a fine map. From the results of linkage analysis we concluded that the fe locus is located in a 2.1±1.0 cM interval from the marker D10mit191 on chromosome 10.
The mouse map location of the fe locus appears to place it in a region homologous to human 6q21. From mapping information obtained from the Mouse Genome Database for Chromosome 10, two genes, Macs and Fyn, were identified as potential candidate genes. But the Macs and Fyn genes were unlikely candidates since Macs and Fyn null mice do not show a faded coat color (Grant et al, 1992; Stumpo et al, 1995). No human candidate gene has yet been localized in this region, which may potentially be the human homology of the fe locus. Thus, the absence of potential human candidate genes renders the characterization of the faded mouse mutation even more pertinent for understanding this disorder.
Although there are several previously described mutations in mice that reduce the intensity of hair pigmentation or cause age-related pigment change (Yokoyama et al, 1990; Fukuta et al, 1991), the descriptions of the coat mutations in all the known mutants seemed to differ from the characteristics and inheritance pattern that we observed in faded mice. Early loss of hair bulb melanocytes has been observed in light (Blt) mice with a mutation at the b-locus (located on chromosome 4) encoding for the melanogenic enzyme TRP-1 (Cable et al, 1993). The phenotype produced by this light mutation involves a defect in mid-anagen phase, resulting in the formation of white hair shafts with pigmented tips. Affected melanocytes exhibit clumping and a reduction in the number of melanosomes, ending in the premature death of all follicular melanocytes when they dislocate from the bulb and incorporate into the hair shaft. This melanotoxicity may be due to toxic intermediates and byproducts of eumelanogenesis because destruction of melanocytes is observed in C57BL mice but not albino mice carrying the allele (Tobin and Paus, 2001). This mutation provided a useful model system for cell death (premature or programmed) in general and for melanocyte death in particular such as is seen in the human disease vitiligo (in which localized melanocyte death occurs) as does the more widespread phenomenon of greying hair (Johnson and Jackson, 1992). Although light (Blt) and faded mice have very similar phenotypes, light (Blt) mice seemed to differ from faded mice with regard to genetic locus and inheritance pattern.
Hair pigmentation is strictly coupled to the anagen stage of the hair cycle (Slominski, 1994). In young animals telogen phase may last 10 days or less, whereas in older mice it may persist for months before a new wave of hair growth begins (Silvers, 1979). Nevertheless, new cycles can be initiated by plucking telogen hairs (Straile et al, 1961). Although faded mice have a normal hair cycle, the hairs of these mice don't have normal melanin content. Therefore, the coat color of homozygote mice becomes progressively lighter with advancing age. In new hair cycles, the hair pigmentation of faded mice display abnormal states. Then the outer fur and under fur become white from the proximal region of hair and show an age-related pigment change toward white. But these shafts appeared normal in structure. Cleared hairs of faded mice display various clumps of pigment granules, which occur sporadically in the medulla of the shaft. Therefore, the phenotype of faded mice may be a reflection of physiological effects related to peculiarities of pigment metabolism or cell death (premature or programmed).
Given the age-related reduction of pigmentation and the number of melanotic melanocytes in hair bulbs, we hypothesize that the gene mutation adversely affects the pigment metabolism of hair bulb melanocytes (e.g. it decrease tyrosinase activity) and cell death (e.g. premature or programmed). Biochemical studies (e.g. tyrosinase activity measurement), electron microscopic examination (e.g. of hair bulb melanocytes) and cloning of the fe gene should elucidate whether these hypotheses are correct.
Acknowledgments
This work was supported by Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0029642).
References
1. Brilliant MH. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res. 2001; 14(2):86–93. PMID: 11310796.
2. Cable J, Jackson IJ, Steel KP. Light (Blt), a mutation that causes melanocyte death, affects stria vascularis function in the mouse inner ear. Pigment Cell Res. 1993; 6:215–225. PMID: 8248019.

3. Edery P, Attie T, Amiel J, Pelet A, Eng C, Hofstra RM, Martelli H, Bidaud C, Munnich A, Lyonnet S. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome). Nat Genet. 1996; 12(4):442–444. PMID: 8630502.

4. Fukuta K, Imamura K, Goto N. Pink-eyed dilution, a coat color mutation in the Japanese field vole (Microtus montebelli). Jikken Dobutsu. 1991; 40(3):375–379. PMID: 1915604.
5. Green EL. Linkage, recombination and mapping. Genetics and probability in animal breeding experiments. 1981. Oxford University Press: New York;p. 77–133.

6. Grant SG, O'Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER. Impaired long-term potentiation, spatial learning and hippocampal development in fyn mutant mice. Science. 1992; 258(5090):1903–1910. PMID: 1361685.
7. Gariepy CE, Williams SC, Richardson JA, Hammer RE, Yanagisawa M. Transgenic expression of the endothelin-B receptor prevents congenital intestinal aganglionosis in a rat model of Hirschsprung disease. J Clin Invest. 1998; 102(6):1092–1101. PMID: 9739043.

8. Johnson R, Jackson IJ. Light is a dominant mouse mutation resulting in premature cell death. Nat Genet. 1992; 1(3):226–229. PMID: 1303241.

9. Kobayashi T, Vicira WD, Pottcrt B, Sakai C, Imokawa G, Hearing VJ. Light Modulation of melanogenic protein expression during the switch from eu- to phaeomelanogenesis. J Cell Sci. 1995; 108:2301–2309. PMID: 7673350.
10. Kunisada T, Yoshida H, Yamazaki H, Miyamoto A, Hemmi H, Nishimura E, Shultz LD, Nishikawa S, Hayashi S. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development. 1998; 125(15):2915–2923. PMID: 9655813.

11. Lamoreux ML. Strain-specific white-spotting patterns in laboratory mice. Pigment Cell Res. 1999; 12(6):383–390. PMID: 10614578.

12. Lehman AL, Silvers WK, Puri N, Wakamatsu K, Ito S, Brilliant MH. The underwhite (uw) locus acts autonomously and reduces the production of melanin. J Invest Dermatol. 2000; 115:601–606. PMID: 10998130.

13. Marks PW, Bandura JL, Shieh DB, Foernzler D, Beier DR, Kwiatkowski DJ. The spontaneous coat color mutant white nose (wn) maps to murine Chromosome 15. Mamm Genome. 1999; 10(7):750–752. PMID: 10384053.
14. Oh YS, Tomita T. Linkage of faded gene (fe) to chromosome 6 of the mouse. Jikken Dobutsu. 1987; 36(1):73–77. PMID: 3816991.
15. Oh YS, Tomita T, Kondo K. Faded, a mutation in the KSB strain of mouse which shows age-related pigment change. Jikken Dobutsu. 1986; 35(2):131–138. PMID: 3732403.

16. O'Reilly FM, Brat DJ, McAlpine BE, Grossniklaus HE, Folpe AL, Arbiser JL. Microphthalmia transcription factor immunohistochemistry: a useful diagnostic marker in the diagnosis and detection of cutaneous melanoma, sentinel lymph node metastases, and extracutaneous melanocytic neoplasms. J Am Acad Dermatol. 2001; 45(3):414–419. PMID: 11511840.
17. Pastural E, Barrat FJ, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, Seger R, Criscelli C, Fischer A, de Saint Basile G. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the Myosin Va gene. Nature Genet. 1997; 16:289–292. PMID: 9207796.
18. Richards KA, Fukai K, Oiso N, Paller AS. A novel KIT mutation results in piebaldism with progressive depigmentation. J Am Acad Dermatol. 2001; 44(2):288–292. PMID: 11174389.

19. Straile WZ, Chase HB, Apenault C. Growth and differentiation of hair follicles between periods of activity and quiescence. J Exp Zool. 1961; 148:205–222. PMID: 13917600.

20. Silvers WK. The coat colors of mice. 1979. New York: Springer-Verlag;p. 1–5.
21. Slominski A, Paus R. Melanogenesis is coupled to murine angen: toward new concepts for the role of melanocytes and the regulation of melanogensis in hair growth. J Invest Dermatol. 1993; 101:90s–97s. PMID: 8326158.
22. Slominski A, Paus R, Plonka P, Chakraborty A, Maurer M, Pruski D, Lukiewicz S. Melanogenesis during the anagen-catagentelogen transformation of the murine hair cycle. J Invest Dermatol. 1994; 102:862–862. PMID: 8006449.

23. Stumpo DJ, Bock CB, Tuttle JS, Blackshear PJ. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc Natl Acad Sci. 1995; 92(4):944–948. PMID: 7862670.

24. Suh JG, Yamanishi T, Matsui K, Tanaka K, Wada K. Mapping of the gracile axonal dystrophy (gad) gene to a region between D5mit197 and D5mit113 on proximal mouse chromosome 5. Genomics. 1995; 27:549–551. PMID: 7558041.
25. Shanske A, Ferreira JC, Leonard JC, Fuller P, Marion RW. Hirschsprung disease in an infant with a contiguous gene syndrome of chromosome 13. Am J Med Genet. 2001; 102(3):231–236. PMID: 11484199.

26. Tomita Y, Miyamura Y, Kono M, Nakamura R, Matsunaga J. Molecular bases of congenital hypopigmentary disorders in humans and oculocutaneous albinism 1 in Japan. Pigment Cell Res. 2000; 13(Suppl 8):130–134. PMID: 11041370.

27. Tobin DJ, Paus R. Graying: gerontobiology of the hair follicle pigmentary unit. Exp Gerontol. 2001; 36(1):29–54. PMID: 11162910.

28. Toyofuku K, Wada I, Valencia JC, Kushimoto T, Ferrans VJ, Hearing VJ. Oculocutaneous albinism types 1 and 3 are ER retention diseases: mutation of tyrosinase or Tyrp1 can affect the processing of both mutant and wild-type proteins. FASEB J. 2001; 15(12):2149–2161. PMID: 11641241.

29. Voisey J, Box NF, van Daal A. A polymorphism study of the human Agouti gene and its association with MC1R. Pigment Cell Res. 2001; 14(4):264–267. PMID: 11549109.

30. Wang C, Kim E, Attaie A, Smith TN, Wilcox ER, Lalwani AK. A PAX3 polymorphism (T315K) in a family exhibiting Waardenburg Syndrome type 2. Mol Cell Probes. 1998; 12(1):55–57. PMID: 9584079.
31. Wada A, Okumoto M, Tsudzuki M. Tawny: a novel light coat color mutation found in a wild population of Mus musculus molossinus, a new allele at the melanocortin 1 receptor (Mc1r) locus. Exp Anim. 1999; 48(2):73–78. PMID: 10374067.
32. Yokoyama T, Silversides DW, Waymire KG, Kwon BS, Takeuchi T, Overbeek PA. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990; 18(24):7293–7298. PMID: 2124349.

Figure 1
Age-related pigment change of affected N2 progenies. A: 1 week, B: 2 weeks, C: 4 weeks, D: 28 weeks.
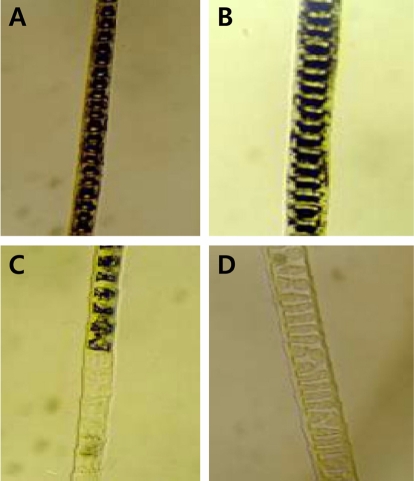
Figure 2
Haplotype analysis of an intraspecific backcross between fe and CBA/J mice [(C57BL/6J-fe/fe×CBA/J)F1×C57BL/6J-fe/fe]. Each column represents a chromosomal haplotype identified in the backcross and inherited from the F1 parent. The number of individuals is listed beneath each column. The black boxes represent fe alleles, and the white boxes represent CBA/J alleles as determined by SSLP scoring with markers indicated at the left.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download