Abstract
Mutations in the transmembrane inner ear (Tmie) gene, which encodes the Tmie protein, have been attributed to deafness autosomal recessive 6 (DFNB6), an autosomal nonsyndromic recessive hearing loss disorder. Although the Tmie gene was identified a few years ago, little is known about subcellular localization of the Tmie protein. In order to address this, we developed a stable cell line expressing Tmie protein. The expression of Myc-tagged Tmie protein was confirmed by Western blot analysis using an anti-Myc antibody and localization of the Tmie protein was confirmed by immunostaining, using the anti-Myc antibody as well as the anti-tmie antibody. Our study demonstrates that the Tmie protein is localized mostly in the cellular membrane and to a lesser extent in cytoplasm. These results suggest that our Tmie expressing stable cell line provides a suitable in vitro model to explore Tmie synthesis and functions.
Deafness is the most common form of sensory impairment in humans. Over the last decade, enormous progress has been made in the discovery of genes involved in hearing and deafness, as well as in the identification of many new loci and specific mutations that cause heritable deafness and vestibular disorders [1]. There has been a particularly tremendous escalation in the localization and identification of genes for nonsyndromic hearing impairment. Inherited deafness in humans is genetically heterogeneous, with effects in any one of more than 100 distinct genes likely to be responsible for nonsyndromic hearing loss [2]. The incidence of hearing loss in humans is high, with a frequency of prelingual deafness as high as 0.1-0.2% and a similar frequency of post-lingual deafness before the third decade of life. Researchers now believe that more than 60% of congenital deafness cases in developed nations are caused by genetic factors [3]. Despite difficulties in analysis of genetically heterogeneous conditions, there has been dramatic progress in the localization and identification of a large number of genes associated with hearing loss during the past several years. The causes of nonsyndromic deafness are complex. Researchers have identified more than 30 genes that, when mutated, may cause nonsyndromic deafness; however, some of these genes have not been fully characterized.
Recently, loss-of-function mutations in the transmembrane inner ear (Tmie) gene have been shown to cause deafness in mice and humans [4-9]. These results indicate that the Tmie gene has a critical role in the auditory system. Previously our research group has reported that circling mice are a possible animal model for deafness. These mice have a 40-kb genomic deletion, including the Tmie gene [6,10-11].
In order to understand the subcellular localization and functional relationships of the Tmie protein, we developed a stable cell line for expressing Tmie protein. The cells (HEK293) were transfected with the Tmie expressing vector [pcDNA 3.1-Myc-His (5.5 Kb)-Tmie] and the expression of Myc-tagged Tmie was confirmed by Western blot analysis and immunostaining analysis using anti-Myc and anti-Tmie antibodies. Our results provide an excellent model for studying the synthesis and localization of Tmie protein.
A Tmie expressing vector [pcDNA 3.1-Myc-His (5.5 Kb)-Tmie] was used [12]. Briefly, mouse Tmie cDNA was amplified by polymerase chain reaction (PCR) using the primer sets: 5'-CTGGACTCTCAGGACCTGCA-3' and 5'-TCAGGAAGCC GCCCTCATTT-3'. The amplified PCR product was ligated into the XhoI and HindIII sites of mammalian expression system vector pcDNA3.1-Myc-His (Invitrogen, Grand Island, NY, USA) to yield the Tmie expression construct pcDNA 3.1-Tmie-Myc-His. DNA sequencing was used to verify the nucleotide sequences of the Tmie expression vector.
The human embryonic kidney cell line (HEK293) was obtained from the American type culture collection. HEK293 was maintained with Dulbecco's modified Eagles medium containing 10% fetal bovine serum, 25 mM HEPES, 100 U/mL, penicillin and 100 µg/mL streptomycin. Cells were cultured at 37℃ at 95% of air and 5% CO2 atmosphere. To generate a stable cell line, we transfected 5 µg of the Tmie expressing vector [pcDNA 3.1-Myc-His (5.5 Kb)-Tmie] which confers neomycin resistance into HEK293 cells, using FuGENE 6 transfection reagent (Roche, Mannheim, Germany) according to manufacturer instruction, Two days after transfection, cells were selected in 500 µg/mL G418 (Duchefa Bioch, Haarlem, Netherlands) for 2 weeks.
Cells (5×106) were lysed in lysis buffer (50 mM Tris-HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1 mM PMSF). Clarified lysates were resolved on 12% SDS-polyacrylamide gels and then transferred to polyvinylidene fluoride transfer membranes (Millipore, Billerica, MA, USA). The membranes were blocked in Tris-buffered saline containing 0.05% Tween 20 and 2% bovine serum albumin for 1 hr at room temperature. Membranes were incubated with an anti-Myc antibody (1:1,000, Cell Signaling Technology, Danvers, MA, USA) for 2 h. Immunoreactive proteins were detected using horseradish peroxidase-conjugated secondary antibody (1:500, Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and enhanced Chemiluminescence reagent (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
For immunofluorescence staining, the stable cells were cultured on glass coverslips in 12-well plates for 24 h in order to detect Tmie localization. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, incubated with antibodies against Myc epitope (1:200), and then incubated with Alexa Flour 633 conjugated goat anti-mouse IgG (Molecular Probes, Grand Island, NY, USA). Nuclei were stained with DAPI. Coverslips were mounted in fluoromount-G (Southern Biotechnology Associates, Birmingham, AL, USA). The mounted samples were scanned with an LSM 510 (Carl Zeiss, Jena, Germany). Several positive clones were expanded and analyzed in this study.
Because we aimed to generate cell lines that stably express Tmie, we transfected the vector containing Tmie and a neomycin resistance gene (Figure 1). In order to monitor expression of Tmie, we introduced a Myc epitope into the end of Tmie gene. The vector sequences were confirmed by an automatic sequencer. Thereafter HEK 293 cells were selected with G418 for stable expression of the transgenes. The Tmie protein expression level of the established cell lines was determined by western blotting analysis as described in methods. The Tmie expression level in the established cell lines was monitored through several passages by Western blot analysis. Stable transgene expression was observed with different levels (Figure 2). No band was detected in untransfected (empty vector) cells.
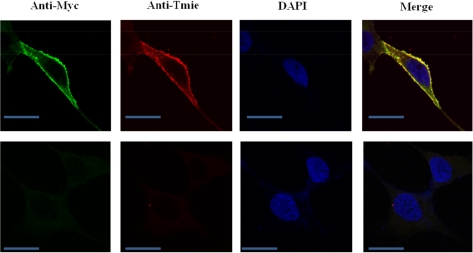
Along with analyzing the expression levels, we also studied the subcellular localization of Tmie in the established cell lines. The subcellular localization of Myc-tagged Tmie was studied by immunofluorescent staining followed by confocal microscopy. Detailed subcellular localization analysis performed with HEK293 cells clearly demonstrated that the Myc-tagged Tmie is expressed predominantly in the plasma membrane (Figure 3).
Inherited hearing loss is genetically heterogeneous and is caused by mutations in gene encoding proteins responsible for a variety of processes, including the maintenance of hair cell structure, neuronal innervations, and trafficking. Recently, a high-throughput detection method of the mutations responsible for hearing loss was established [13]. It has been found that Tmie is responsible for nonsyndromic hearing loss in humans [7-9]. The Tmie protein has a signal peptide and at least one transmembrane domain [5]. According to the proposed structure of Tmie, the mature protein may be localized in the plasma membrane and serve as a site of interaction for other molecules through its highly charged C-terminal domain. Several papers suggest that the Tmie protein may require maturation of sensory cells as well as the normal development of the maintenance of stereocilia [5,14-16].
To examine the subcellular localization of the Tmie protein we generated a cell line that expresses the Tmie protein. The Tmie protein expression in HEK293-Tmie cells was monitored by using immunostaining analysis and the localization was observed through confocal microscopy. From this study we concluded that the Tmie protein is mostly present in the cellular membrane, which suggests that Tmie protein may play an important role in signal trafficking between the cells or in signal transduction in the auditory system. Tmie protein probably joins with other proteins while transducting signals in the auditory system.
We predict that the Tmie protein may play an important functional role in cell metabolism and signal transduction. However, more specific study is needed to understand its functional role in adjacent cells. It remains to be determined what specific role Tmie plays in hair cell development and specifically, what advantages its localization and membrane association provide. However the data provided in this study offer a solid foundation from which these questions can better be addressed.
In summary, we established a Tmie-expressing cell line on HEK 293 cells, and the expression of Tmie was confirmed by both western blotting and confocal microscopy. Considering the shortcomings involved in functional analysis of Tmie, our method ensures stable and high expression of the protein, thus providing an excellent model for further research into its biological effects. Further examination of the impact of Tmie and its protein will provide insight into the pathways that are critical for normal maturation of sensory cell function. Analysis of Tmie expression and localization will also help to define the molecular mechanism of hearing loss in humans with defects at the DFNB6 locus, and will provide a system to investigate potential therapies that rescue sensory cells and preserve auditory function.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (KRF-2007-521-E00140).
References
1. Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010; 68(2):293–308. PMID: 20955936.

2. Raviv D, Dror AA, Avraham KB. Hearing loss: a common disorder caused by many rare alleles. Ann N Y Acad Sci. 2010; 1214:168–179. PMID: 21175685.

3. Lalwani AK, Castelein CM. Cracking the Auditory Genetic Code: nonsyndromic hereditary hearing impairment. Am J Otol. 1999; 20:115–132. PMID: 9918184.
4. Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, Ramesh A, Srisailpathy S, Deshmukh D, Riazuddi S, Griffith AJ, Friedman TB, Smith RJ, Wilcox ER. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet. 2002; 71(3):632–636. PMID: 12145746.

5. Mitchem KL, Hibbard E, Beyer LA, Bosom K, Dootz GA, Dolan DF, Johnson KR, Raphael Y, Kohrman DC. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet. 2002; 11(16):1887–1898. PMID: 12140191.

6. Cho KI, Suh JG, Lee JW, Hong SH, Kang TC, Oh YS, Ryoo ZY. The circling mouse (C57BL/6J-cir) has a 40-kilobase genomic deletion that includes the transmembrane inner ear (tmie) gene. Comp Med. 2006; 56:476–481. PMID: 17219777.
7. Santos RL, El-Shanti H, Sikandar S, Lee K, Bhatti A, Yan K, Chahrour MH, McArthur N, Pham TL, Mahasneh AA, Ahmad W, Leal SM. Novel sequence variants in the TMIE gene in families with autosomal recessive nonsyndromic hearing impairment. J Mol Med. 2006; 84(3):226–231. PMID: 16389551.

8. Sirmaci A, Oztükmen-Akay H, Erbek S, Incesulu A, Duman D, Taşir-Yilmaz S, Ozdağ H, Tekin M. A founder TMIE mutation is a frequent cause of hearing loss in Southeastern Anatolia. Clin Genet. 2009; 75(6):562–567. PMID: 19438934.
9. Yang JJ, Su MC, Chien KH, Hsin CH, Li SY. Identification of novel variants in the TMIE gene of patients with nonsyndromic hearing loss. Int J Pediatr Otorhinolaryngol. 2010; 74(5):489–493. PMID: 20206386.

10. Lee JW, Ryoo ZY, Lee EJ, Hong SH, Chung WH, Lee HT, Chung KS, Kim TY, Oh YS, Suh JG. Circling mouse, a spontaneous mutant in the inner ear. Exp Anim. 2002; 51:167–171. PMID: 12012726.

11. Cho KI, Lee JW, Kim KS, Lee EJ, Suh JG, Lee HJ, Kim HT, Hong SH, Chung WH, Chang KT, Hyun BH, Oh YS, Ryoo ZY. Fine mapping of the circling (cir) gene on the distal portion of mouse chromosome 9. Comp Med. 2003; 53:642–648. PMID: 14727813.
12. Nam YY, Karuppasamy S, Byoungkwon Park, Kwon HJ, Suh JG. Production and Characterization of polyclonal antibody to transmembrane inner ear protein. Lab Anim Res. 2009; 25(2):145–150.
13. Kothiyal P, Cox S, Ebert J, Husami A, Kenna MA, Greinwald JH, Aronow BJ, Rehm HL. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010; 10:10. PMID: 20146813.

14. Chung WH, Kim KR, Cho YS, Cho DY, Woo JH, Ryoo ZY, Cho KI, Hong SH. Cochlear pathology of the circling mouse: a new mouse model of DFNB6. Acta Otolaryngol. 2007; 127(3):244–251. PMID: 17364360.

15. Shin MJ, Lee JH, Yu DH, Kim HJ, Bae KB, Yuh HS, Kim MO, Hyun BH, Lee S, Park R, Ryoo ZY. Spatiotemporal expression of tmie in the inner ear of rats during postnatal development. Comp Med. 2010; 60(4):288–294. PMID: 20819378.
16. Gleason MR, Nagiel A, Jamet S, Vologodskaia M, López-Schier H, Hudspeth AJ. The transmembrane inner ear (Tmie) protein is essential for normal hearing and balance in the zebrafish. Proc Natl Acad Sci USA. 2009; 106(50):21347–21352. PMID: 19934034.

Figure 1
The map and sequences of the expressing vector containing the Tmie gene (pcDNA 3.1-Myc-His).
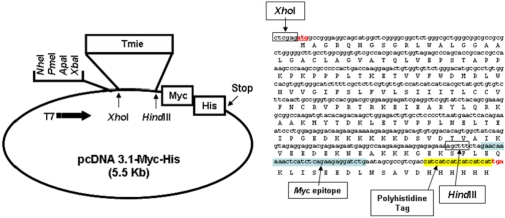
Figure 2
Tmie expression in Tmie-HEK293 cell lines. Tmie (~21 kDa) expression was detected by western blotting using an anti-Myc antibody. Lane 1: lysate from HEK293 cells transfected expressing an empty vector, Lane 2: lysate from HEK293 cells transfected expressing a vector containing the Tmie gene.
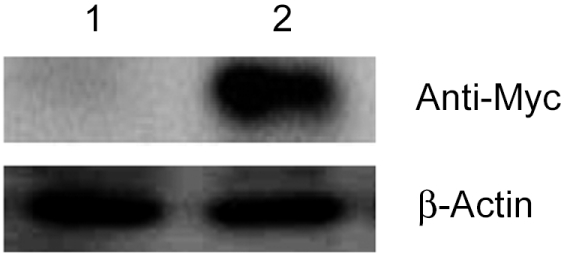
Figure 3
Subcellular localization of Tmie in Tmie-expressing cells (Tmie-HEK293). Expression of the Tmie protein was detected in the stable cell line by confocal microscopy. Tmie protein expression was found in the cellular membrane (red) and to a lesser extent in cytoplasm. Upper panels are expressing vector-treated cells. Lower panels are mock-treated cells. Merge indicates merged images of Myc (green) and Tmie (red) immunofluorescence. (Scale bar=20 µm).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download