Abstract
Eimeria tenella and Eimeria maxima are important pathogens causing intracellular protozoa infections in laboratory avian animals and are known to affect experimental results obtained from contaminated animals. This study aimed to find a fast, sensitive, and efficient protocol for the molecular identification of E. tenella and E. maxima in experimental samples using chickens as laboratory avian animals. DNA was extracted from fecal samples collected from chickens and polymerase chain reaction (PCR) analysis was employed to detect E. tenella and E. maxima from the extracted DNA. The target nucleic acid fragments were specifically amplified by PCR. Feces secreting E. tenella and E. maxima were detected by a positive PCR reaction. In this study, we were able to successfully detect E. tenella and E. maxima using the molecular diagnostic method of PCR. As such, we recommended PCR for monitoring E. tenella and E. maxima in laboratory avian facilities.
Coccidiosis induced by Eimeria species infection is an important parasitic disease of poultry [1]. Losses include mortality, morbidity, and cost of preventative or therapeutic drugs and/or vaccinations. In addition, many of the in-feed medications commonly used for prevention of infections with Eimeria species have become less effective as some strains of parasites have developed reduced susceptibility to anticoccidials [2]. This development suggests that coccidiosis is likely to have a greater impact on the profitability of broiler meat production in the future [2]. Diagnosis of coccidiosis is based on comparing clinical features, gut pathology in the host, parasite properties such as morphology of different parasite stages in fecal material or intestine, and the prepatent period [3,4]. This avenue is not only very subjective and time-consuming, but is also unreliable since different species have overlapping properties [5]. Consequently, the development of molecular tools for identification and characterization of these parasites is important. Several polymerase chain reaction (PCR)-based assays targeting different regions of the Eimeria genome have been described, including 5S rRNA [6], small subunit rRNA [6,7], sporozoite antigen gene EASZ240/160 [8], internal transcribed spacer-1 (ITS-1) [9-12], and ITS-2 [13-15]. Nevertheless, the practical implementation of these methods in routine diagnostic and epidemiological studies of chicken coccidiosis [9,12] has been limited, and as such the assays are still regarded as experimental.
The most commonly used protocol for identification of Eimeria species is based on morphological observation of oocysts under a microscope. This procedure is both labor- and time-consuming. The bottleneck for an effective molecular diagnostic procedure is not the PCR amplification of genomic coccidial DNA, since that has been shown to be highly sensitive, but rather the preparation of the DNA from oocysts [16]. The oocyst wall of avian coccidia is particularly rigid and resistant to chemical and mechanical forces [2,17]. A number of methods to rupture the coccidial oocyst wall have been suggested [18-20], but the most widespread technique is to add glass beads to the oocyst suspension and then vortex until the glass-bead grinding ruptures the oocysts [8,16,21-23].
E. tenella and E. maxima are important pathogens known to cause avian coccidiosis in laboratory avian animals and to influence experimental results obtained from contaminated animals [1,24]. This study aimed to find a fast, sensitive, and efficient protocol for the molecular identification of these important protozoan pathogens, in laboratory avian facilities.
This research was conducted using three-day-old chickens (n=10) at the animal facility of the Center for Animal Resources Development, Wonkwang University, Korea. Animals were acclimatized and kept in an animal facility room with regulated temperature (28±2℃), humidity (50±5%), and light/dark cycle (12/12 h). The chickens were fed commercial post-broiler diet without antibiotics and coccidiostat (Hanil Feed Co, Yongin, Korea).Tap water was available ad libitum. The chickens were kept in wire-floored grower cages throughout the study period. All studies were performed in accordance with the Guide for Animal Experimentation and approved by the Institutional Animal Care and Use Committee of Wonkwang University. All efforts were made to minimize pain or discomfort to study animals.
E. tenella and maxima were kindly provided by professor Wongi Min at Gyeongsang National University in Korea. E. tenella and E. maxima were cleaned by flotation with 5.25% sodium hypochlorite and washed three times with phosphate buffered saline. Chickens were treated orally by gavage using a 24-gauge, stainless steel animal feeding tube (Popper & Sons, New York, USA) attached to a 3-mL syringe. The oral infectious dose of approximately 104 oocysts each of E. tenella and E. maxima in 1 mL of saline was administered. Fecal materials were collected from 6 to 10 days post-infection and analyzed for the presence of coccidial oocysts using a standard fecal flotation technique [25]. Briefly, 5 mL from each sample was pelleted by centrifugation at 1,500 g for 5 min. The resulting pellet was resuspended in saturated sodium chloride (aqueous) and passed through a 1-mm mesh sieve to remove coarse fecal debris. The resulting filtrate was used for standard gravity vial fecal flotation using 22 mm×22 mm coverslips. After flotation, the coverslip was mounted on a slide and examined in its entirety for the presence of coccidial oocysts. For oocyst-positive samples, the remaining 45 mL of the intestinal tract contents were incubated at 26℃ for seven days to permit completion of sporulation. Sporulated oocysts were washed to remove potassium dichromate, concentrated by salt flotation, resuspended in distilled water, and then stored at 4℃ until DNA isolation.
For DNA extraction, each sample was cleaned with sodium hypochlorite solution (5-6% active chlorine) for 10 min at 4℃, washed three times with deionized water and resuspended in extraction buffer (10 mM Tris-Cl, 50 mM EDTA, pH 8.0). The oocysts and sporocysts were completely broken down by vortexing with half the volume of 425-600 µm acid-washed glass beads (Sigma-Aldrich, St. Louis, MO, USA). The lysate was centrifuged at 14,000 g for 10 min to eliminate debris and then digested with DNAse-free RNAse A (20 µg/mL) at 37℃ for 1 h. A further digestion with proteinase K (100 µg/mL) and sodium dodecyl sulfate (SDS, 0.5%) was performed at 50℃ for 2 h. The DNA was then extracted once with one volume of phenol, phenol/chloroform, and chloroform, and precipitated with ethanol and ammonium acetate. The pellet was washed with 70% ethanol. The DNA was eluted in Tris-EDTA buffer (pH 8.0), and an aliquot was used for PCR amplification. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower™ PCR PreMix; Bioneer, Daejeon, Korea) containing 2.5 U of Taq DNA polymerase, 250 µM each of deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and the gel loading dye. The volume was adjusted with distilled water to 20 µL. The primers of E. tenella were amplified from genomic DNA using a species-specific forward primer (5'-AATTTAGTCCATCGCAACCCTTG-3') and reverse primer (5'-CGAGCGCTCTGCATACGACA-3'). The primers of E. maxima were amplified from genomic DNA using a species-specific forward primer (5'-GTGGGACTGTGGTGATGGGG-3') and reverse primer (5'-ACCAGCATGCGCTCACAACCC-3'). The reaction mixtures were subjected to denaturation at 96℃ for 5 min followed by 35 cycles of denaturation at 95℃ for 30 sec, annealing at 58 or 65℃ for 30 sec, and extension at 72℃ for 1 min, and a final extension step of 72℃ for 3 min. Samples were kept at 4℃ until analyzed. Reactions were conducted using My Genie 32 Thermal Block PCR (Bioneer). Each sample (10 µL) was analyzed by electrophoresis on 2% agarose gels stained with 0.5 µg/mL ethidium bromide. Necropsies were conducted and the intestines were submitted for gross examination and trimming. The trimmed tissue was fixed in 10% neutral buffered formalin, and embedded in paraffin. Four µm sections were made and stained with hematoxylin and eosin for histopathological examination.
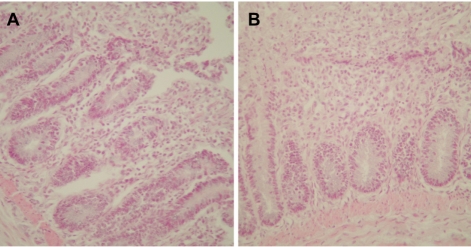
A PCR assay, based on the amplification of internal transcribed spacer 1 (ITS1) regions of ribosomal DNA, was developed for the chicken coccidian species E. tenella and E. maxima. E. tenella was successfully identified by a distinct 278 base pair (bp) band on agarose gels (Figure 1). Also, amplicons were obtained for 205 bp by species-specific PCR for E. maxima. Positive PCR reactions were obtained with fecal samples (Figure 2). Histopathologic findings of the Eimeria-infected intestines revealed severe villous destruction and large numbers of coccidia oocysts in villous epithelial cells (Figure 3).
Coccidiosis of domestic fowl is a worldwide disease caused by obligatory intracellular protozoa of the genus Eimeria. The disease is responsible for important economic losses in poultry production. E. tenella and E. maxima are important pathogens causing avian coccidiosis in laboratory avian animals and known to affect experimental results obtained from contaminated animals [1,24]. The disease is characterized by enteric lesions of variable extent and severity, which reduce the absorptive function of the intestinal mucosa, thus leading to weight loss, diarrhea, poorer feed conversion, and a higher mortality in affected flocks [26]. Due to the thick and resistant oocyst wall of Eimeria species [17] several means of breaking down the oocyst wall have been described, including sonication [26], hot phenol incubation [6,27], repeated freezing and thawing [19], enzyme digestion after sodium hypochlorite incubation [27], passage through a high pressure cell [18], grinding in liquid nitrogen [7,19] and grinding by glass beads [16,21,23]. The use of glass beads is effective and is the most commonly used procedure, although reported bead sizes and grinding times may differ. The grinding efficacy depends on oocyst contact with the glass beads, the container wall, and/or each other. When the oocyst concentration is low, the oocyst grinding may be less efficient and thus a limiting factor. Another limiting factor may be the amount of fecal remains in a sample. When heavy burdens of fecal remains are present, the final fraction of oocysts requires a longer time to grind, possible due to interference by the fecal debris. Consequently, the yield of DNA may not be directly proportional to the amount of oocysts. In a practical situation the oocysts in field material are isolated by the saturated sodium chloride flotation technique [28]. Our study shows that depending on the quality of the sample, repeated flotation might be necessary to obtain a sufficiently clean test sample to prevent fecal inhibition. Traditional methods for species differentiation of chicken Eimeria species are not only time-consuming, cumbersome, and labor intensive, but they are insensitive and highly subjective. The advantage of using PCR compared to traditional methods is its high sensitivity and, if optimal, objectivity, making it ideal for applied parasitology such as diagnostics, prevention, and control of disease.
In this study, the PCR analysis used as a molecular diagnostic method was able to successfully detect Eimeria species. Thus, PCR is recommended for monitoring E. tenella and E. maxima in laboratory animals with avian coccidiosis.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2010-0021940).
References
1. Dalloul RA, Lillehoj HS. Poultry coccidiosis: recent advancements in control measures and vaccine development. Expert Rev Vaccines. 2006; 5(1):143–163. PMID: 16451116.

2. Wilson PA, Fairbairnz D. Biochemistry of sporulation in oocysts of Eimeria acervulina. J Protozool. 1961; 8(4):410–416.
3. Long PL, Millard BJ, Joyner LP, Norton CC. A guide to laboratory techniques used in the study and diagnosis of avian coccidiosis. Folia Vet Lat. 1976; 6(3):201–217. PMID: 1010500.
4. Long PL, Reid WM. A guide for the diagnosis of coccidiosis in chickens. Research report. 1982. 404:The University of Georgia, College of Agriculture Experiment Stations;p. 1–17.
5. Long PL, Joyner LP. Problems in identification of species of Eimeria. J Protozool. 1984; 31(4):535–541. PMID: 6392531.
6. Stucki U, Braun R, Roditi I. Eimeria tenella: characterization of a 5S ribosomal RNA repeat unit and its use as a species-specific probe. Exp Parasitol. 1993; 76(1):68–75. PMID: 8467900.

7. Tsuji N, Ohta M, Kawazu S, Kamio T, Isobe T, Shimura K, Fujisaki K. DNA polymorphism of srRNA gene among Eimeria tenella strains isolated in Japan. J Vet Med Sci. 1999; 61(12):1331–1333. PMID: 10651056.

8. Molloy JB, Eaves FW, Jeston PJ, Minchin CM, Stewart NP, Lew AE, Jorgensen WK. Detection of Eimeria acervulina using the polymerase chain reaction. Avian Dis. 1998; 42(1):119–123. PMID: 9533088.
9. Lew AE, Anderson GR, Minchin CM, Jeston PJ, Jorgensen WK. Inter- and intra-strain variation and PCR detection of the internal transcribed spacer 1 (ITS-1) sequences of Australian isolates of Eimeria species from chickens. Vet Parasitol. 2003; 112(1-2):33–50. PMID: 12581583.

10. Schnitzler BE, Thebo PL, Mattsson JG, Tomley FM, Shirley MW. Development of a diagnostic PCR assay for the detection and discrimination of four pathogenic Eimeria species of the chicken. Avian Pathol. 1998; 27(5):490–497. PMID: 18484033.
11. Schnitzler BE, Thebo PL, Tomley FM, Uggla A, Shirley MW. PCR identification of chicken Eimeria: a simplified read-out. Avian Pathol. 1999; 28(1):89–93. PMID: 16147553.

12. Su YC, Fei AC, Tsai FM. Differential diagnosis of five avian Eimeria species by polymerase chain reaction using primers derived from the internal transcribed spacer 1 (ITS-1) sequence. Vet Parasitol. 2003; 117(3):221–227. PMID: 14630430.

13. Gasser RB, Woods WG, Wood JM, Ashdown L, Richards G, Whithear KG. Automated, fluorescence-based approach for the specific diagnosis of chicken coccidiosis. Electrophoresis. 2001; 22(16):3546–3550. PMID: 11669540.

14. Lien YY, Sheu SC, Liu HJ, Chen SC, Tsai MY, Luo SC, Wu KC, Liu SS, Su HY. Cloning and nucleotide sequencing of the second internal transcribed spacer of ribosomal DNA for three species of Eimeria from chickens in Taiwan. Vet J. 2007; 173(1):184–189. PMID: 16314128.

15. Woods WG, Whithear KG, Richards DG, Anderson GR, Jorgensen WK, Gasser RB. Single-strand restriction fragment length polymorphism analysis of the second internal transcribed spacer (ribosomal DNA) for six species of Eimeria from chickens in Australia. Int J Parasitol. 2000; 30(9):1019–1023. PMID: 10980293.

16. Fernandez S, Pagotto AH, Furtado MM, Katsuyama AM, Madeira AM, Gruber A. A multiplex PCR assay for the simultaneous detection and discrimination of the seven Eimeria species that infect domestic fowl. Parasitology. 2003; 127(Pt 4):317–325. PMID: 14636018.
17. Ryley JF. Hammond DM, editor. Cytochemistry, physiology, and biochemistry. The Coccidia. 1973. Baltimore: University Park Press;p. 151–154.
18. Abrahamsen MS, Clark TG, White MW. An improved method for isolating RNA from coccidia n oocysts. J Parasitol. 1995; 81(1):107–109. PMID: 7876962.
19. Jinneman KC, Wetherington JH, Hill WE, Adams AM, Johnson JM, Tenge BJ, Dang NL, Manger RL, Wekell MM. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. oocysts directly from raspberries. J Food Prot. 1998; 61(11):1497–1503. PMID: 9829192.

20. Nakamura T, Konishi T, Kawaguchi H. Isoenzymes of chicken coccidia in Japan. Nihon Juigaku Zasshi. 1986; 48:587–590. PMID: 3735889.
21. MacPherson JM, Gajadhar AA. Differentiation of seven Eimeria species by random amplified polymorphic DNA. Vet Parasitol. 1993; 45(3-4):257–266. PMID: 8447068.

22. Procunier JD, Fernando MA, Barta JR. Species and strain differentiation of Eimeria spp. of the domestic fowl using DNA polymorphisms amplified by arbitrary primers. Parasitol Res. 1993; 79(2):98–102. PMID: 8475039.
23. Shirley MW. Enzyme variation in Eimeria species of the chicken. Parasitology. 1975; 71(3):369–376. PMID: 1202411.

24. McDougald LR. Saif YM, editor. Protozoal infections. Diseases of Poultry. 2003. Ames: Iowa State Press;p. 973–991.

25. Reid WM, Long PL. A diagnostic chart for nine species of fowl coccidian. Univ Ga Coll Agric Res Rep. 1979; 335:1–24.
26. Stotish RL, Wang CC, Meyenhofer M. Structure and composition of the oocyst wall of Eimeria tenella. J Parasitol. 1978; 64(6):1074–1081. PMID: 739302.

27. Zhao X, Duszynski DW, Loker ES. A simple method of DNA extraction for Eimeria species. J Microbiol Methods. 2001; 44(2):131–137. PMID: 11165342.

28. Shirley MV. Eckert J, editor. Eimeria species and strains. COST89/820, Biotechnology, Guidelines on Techniques in Coccidiosis Research. 1995. Luxembourg: European Commission;p. 1–25.
Figure 1
Amplification of sample DNA by species-specific polymerase chain reaction (PCR) for Eimeria tenella was identified using 2% agarose gel electrophoresis. Lane P: positive control, N: negative control, T1-T5: DNA extracted from fecal samples of E. tenella-infected chickens.
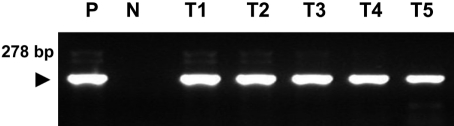
Figure 2
Amplification of sample DNA by species-specific PCR for Eimeria maxima was identified using 2% agarose gel electrophoresis. Lane P: positive control, N: negative control, M1-M5: DNA extracted from fecal samples of E. maxima-infected chickens.
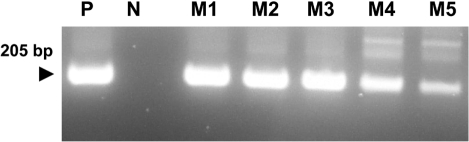
Figure 3
Histopathologic findings from Eimeria-infected chicken intestines revealing severe villous destruction and large numbers of coccidia oocysts in villous epithelial cells. (A) Intestine infected with Eimeria tenella. (B) Intestine infected with Eimeria maxima. Hematoxylin and eosin stain, ×400.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download