Abstract
Reptiles are used for various purposes these days, including public exhibits, medicinal applications, and as laboratory animals. As the international exchange of reptiles has gradually increased, more people have had the opportunity to come in contact with these animals. Snakes typically live in the rhizosphere where various bacterial strains exist and as such they can lead to opportunistic human diseases. When snakes are encountered in veterinary medicine, it is necessary to monitor their microflora. Native microflora of reptiles imported from other countries has not yet been reported in Korea. In this study, oral and cloacae samples were collected from 18 Burmese pythons transported from Vietnam. The specimens were incubated at 37℃ for 18 h to produce colony growth under aerobic condition and isolated colonies were then identified using a VITEK automated identification system. There were fourteen types of aerobic bacteria isolated from both oral and cloacae samples, nine from only oral specimens, and fifteen from only cloacae specimens. Most bacteria isolated were opportunistic pathogens of humans which therefore have the potential to induce disease in people. Based on the microflora and the prevalence of bacterial strains in snakes, quarantine procedures for reptiles transported internationally should be strengthened. Characterization of the microflora of reptiles with the potential to induce zoonosis should be performed in those used as laboratory animals and to prevent zoonotic outbreaks in the general population as well as among veterinarians.
Recently studies using reptiles have increased in the areas of infectious disease, comparative anatomical physiology, the evaluation of phylogenic relations with birds and other vertebrates, stem cell experiments, and therapeutic drug development [1-3]. Various peptides isolated from snake venom have been used for multiple purposes. For example, 1-amino acid oxidase from Bothrops leucurus venom has been studied for inhibition of platelet aggregation [4], and cathelicidin-BF (BF-30) from Bungarus fasciatus has potency to inhibit bacterial proliferation [5]. As medicinal applications of venom increase, its risk to human health should be examined.
Even though snake health can be affected by bacteria, there have not been many studies of the distribution of bacteria in snakes or the influence of predominant bacteria in snakes. For example, gram-positive bacteria such as Corynebacterium and Staphylococcus are predominant in the oral cavities of healthy snakes but gram-negative bacteria, such as Pseudomonas aeruginosa, Providencia rettgeri, and Pseudomonas maltophilia are predominant in the oral cavities of snakes with stomatitis [6]. Some bacteria not only affect snakes but also affect humans [7-8].
Enterococcus faecalis is normal flora in the gastrointestinal tract, but may gain resistance towards antibiotics during pathological conditions [9]. Enterobacter is known to cause urinary and respiratory diseases in humans [6]. Proteus sp. which can induce urinary disease often causes nosocomial outbreaks [10]. Pseudomonas sp. can breakdown non-specific host defenses [6]. Although Stenotrophomonas maltophilia is commonly found in the environment, it can induce human diseases including endocarditis, sepsis, meningitis, peritonitis, and soft tissue and wound infections [11].
The purpose of this study was to identify the aerobic bacteria of internationally transported snakes, investigate the microflora of those used as experimental animals, and ascertain the opportunistic pathogens among the characterized bacteria, in an effort to establish the need for improved inspections related to the international trade of wild animals, including snakes.
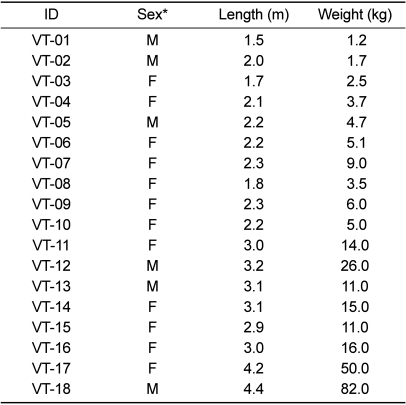
Samples for bacteria cultures were collected from 18 Burmese pythons (Python molurus bivittatus) transported from Anh Thien's farm located in Ho Chi Minh, Vietnam to a private zoo in Korea on May 17, 2007. A total of six males and twelve female snakes which had been maintained in a natural state at the farm were caught and transported to Korea. Before moving to the zoo, samples were collected from the snakes in the Inchon airport cargo area. The snakes were distinguished by their individual characteristics and renamed VT-01 to VT-18 (Table 1). All animals were clinically described as normal. The largest of the snakes reached a length of 4.4 m and a body weight of 82 kg, which in fact was the largest snake ever transported to Korea. The body size of the smallest snake was 1.5 m and 1.24 kg. Animals were immobilized during sample collection. To obtain oral samples, a sterile transport swab with liquid Amies Medium (Copan Innovation, Brescia, Italy) was introduced between the larynx and the bottom of the inner gum line. Cloacae samples were obtained in a similar fashion. The methods of sample collection and storage were not designed to support recovery of anaerobic bacteria. Samples were inoculated on culture plates with 5% sheep blood agar (Becton, Dickinson and Company, NJ, USA) and incubated at 37℃ for 18 hours to produce colony growth under aerobic conditions.
Standard procedures were used for bacterial isolate identification, including Gram stain reaction and colony morphology [12]. Presumptive identification was carried out for differential identification as follows:
Triple sugar iron (TSI) agar was used for the differentiation of Enterobacteriaceae cultivated on selective or moderately selective media on the basis of lactose, glucose, and sucrose fermentation, and the production of hydrogen sulfide (H2S) and other gases. To perform the test, a test tube of TSI media was inoculated by stabbing the butt with a needle which had touched the surface of the center of a colony and a slope was then streaked. After 18-48 h of incubation, the tube was examined for the following: only acid, or acid and gas in the butt; acid or alkaline or no change in the slope; and H2S production. Acidification was detected as yellow-colored gas production by the formation of small bubbles in the agar, and H2S production by blackening of the medium.
Indole tests were also performed which screen for the ability of an organism to degrade the amino acid tryptophan and produce indole. A tube of tryptone broth was inoculated with a small amount of pure culture and incubated at 35℃ for 24 to 48 h. To test for indole production, five drops of Kovac's reagent were added directly to the tube. A positive indole test was indicated by the formation of a pink to red color ("cherry-red ring") in the reagent layer on top of the medium within seconds of adding the reagent. If a culture was indole negative, the reagent layer remained yellow or became slightly cloudy.
Citrate utilization tests were used to identify bacteria which utilize as one of their starting products of metabolism a compound called citrate which is the ionized form of citric acid. Bacterial colonies were picked up with a straight wire and inoculated into a slope of Simmons' citrate agar and incubated for 24 h at 35℃. As citrate was utilized from the media by the bacteria, the pH of the medium changed. As the pH became more alkaline, the media changed from green to blue (alkaline). A green color represented a negative result, and blue denoted a positive state of growth [13].
After cultivating at 35℃ for 18-24 h, gram negative bacterial samples were analyzed using an API 20NE System (bioMérieux, Craponne, France) and gram positive bacterial samples were analyzed using a BBL Crystal Identification System (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). To confirm these results, the isolated colonies were reanalyzed using a VITEK II System (bioMérieux) and standard biochemical assays were used for identification of bacterial species. The isolated colonies were incubated under standard conditions, i.e., gram positive bacteria for 8-24 h, gram negative bacteria for 18-24 h. An organism suspension was made in 0.45-0.5% sterile saline (0.9% NaCl, pH 5.5-7.2) from a pure culture in a clear plastic polystyrene tube and standardized to McFarland 0.5 standard using a DensiCHEK turbidity meter. Readings were performed every 15 min.
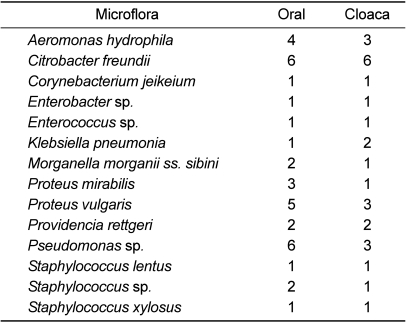
Fourteen bacterial strains, including Aeromonas hydrophila, Citrobacter freundii, Corynebacterium jeikeium, Enterobacter sp., Enterococcus sp., Klebsiella pneumoniae, Morganella morganii ss. sibini, Proteus mirabilis, Proteus vulgaris, Providencia rettgeri, Pseudomonas species, Staphylococcus lentus, Staphylococcus sp., and Staphylococcus xylosus were isolated from both oral and cloaca samples (Table 2). The incidence rate of Citrobacter freundii (33%) in both oral and cloaca cultures was higher than that of the other strains. The incidence rate of Pseudomonas sp. in oral samples (33%) was much higher than in cloaca samples (17%). Most incidence rates in oral samples were higher than in cloaca samples. The incidence rates in oral samples from Proteus vulgaris, Aeromonas hydrophila, Proteus mirabilis, Morganella morganii ss. sibini, and Providencia rettgeri were 28, 22, 17, 11, and 11%, respectively (Table 3), and those in cloaca samples from Proteus vulgaris, Aeromonas hydrophila, Proteus mirabilis, Morganella morganii ss. sibini, and Providencia rettgeri were 17, 17, 6, 6, and 11%, respectively (Table 4).
All isolates were opportunistic pathogens of humans, especially more dangerous to immunosuppressed individuals such as organ transplanted patients.
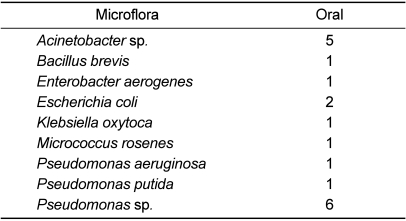
Nine bacterial strains, including Acinetobacter species, Bacillus brevis, Enterobacter aerogenes, Escherichia coli, Klebsiella oxytoca, Micrococcus rosenes, Pseudomonas aeruginosa, Pseudomonas putida, and Pseudomonas species were isolated from oral samples.
Pseudomonas species (33%) were more frequently found in oral cultures compared to the others. The next most frequent oral isolate was Acinetobacter species (28%). All oral isolates were opportunistic pathogens of humans except for Acinetobacter species.
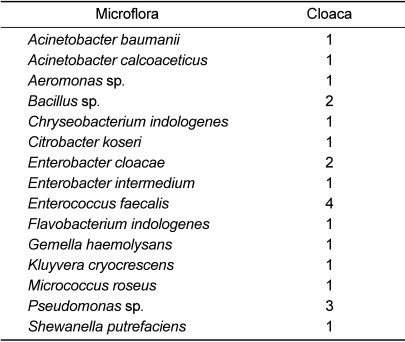
Fifteen bacterial strains, including Acinetobacter baumanii, Acinetobacter calcoaceticus, Aeromonas sp., Bacillus sp., Chryseobacterium indologenes, Citrobacter koseri, Enterobacter cloacae, Enterobacter intermedium, Enterococcus faecalis, Flavobacterium indologenes, Gemella haemolysans, Kluyvera cryocrescens, Micrococcus roseus, Pseudomonas sp., and Shewanella putrefaciens were isolated from cloaca samples.
The characterized bacteria in cloaca samples varied in comparison to those in oral samples. The incidence of Enterococcus faecalis was four times higher than others. The incidence rates of Pseudomonas sp., Bacillus sp., and Enterobacter cloacae were 11% or more in cloaca samples (17, 11, and 11%, respectively). All isolates identified were opportunistic pathogen of humans.
Fourteen bacterial strains (Aeromonas hydrophila, Citrobacter freundii, Corynebacterium jeikeium, Enterobacter sp., Enterococcus sp., Klebsiella pneumonia, Morganella morganii ss. sibini, Proteus mirabilis, Proteus vulgaris, Providencia rettgeri, Pseudomonas sp., Staphylococcus lentus, Staphylococcus sp., and Staphylococcus xylosus) were isolated from both oral and cloaca samples. Nine bacterial strains (Acinetobacter species, Bacillus brevis, Enterobacter aerogenes, Escherichia coli, Klebsiella oxytoca, Micrococcus rosenes, Pseudomonas aeruginosa, Pseudomonas putida, and Pseudomonas sp.) were isolated from oral samples. Fifteen bacterial strains (Acinetobacter baumanii, Acinetobacter calcoaceticus, Aeromonas sp., Bacillus sp., Chryseobacterium indologenes, Citrobacter koseri, Enterobacter cloacae, Enterobacter intermedium, Enterococcus faecalis, Flavobacterium indologenes, Gemella haemolysans, Kluyvera cryocrescens, Micrococcus roseus, Pseudomonas sp., and Shewanella putrefaciens) were isolated from cloaca samples. All isolates identified were opportunistic pathogen of humans [14-22].
These results suggest that individuals involved in international snake transportation or studies using these animals run the risk of being infected by opportunistic pathogen during inspection, transport, or conducting research with them. The possibility of infection is especially high for persons who are sick or immunocompromised.
Snake species are estimated at 3,315 as of Jan, 2011 [23] and most live in the subterranean region. There are also many opportunistic pathogens such as Burkholderia, Ochrobactrum, and Stenotrophomonas in the region [24]. Therefore, it stands to reason that snakes may carry a number of zoonotic pathogens. Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) is a non-fermenting gram-negative bacillus, associated with infections such as endocarditis, sepsis, meningitis, peritonitis, and soft tissue and wound infections [11], which is intrinsically resistant to broad-spectrum antibiotics [25], and has an increased potential to induce disease in humans who have malignancies, chronic respiratory diseases, and endotracheal intubation [26]. In addition to secondary infections arising from bacteria coming from the snake's mouth inoculated at the time of a bite, aerobic bacteria are also responsible for snakebite-associated wound infections [27].
Wild-caught snakes are a popular and traditional food; therefore, it is very likely that people who consume reptile meat are in danger of contracting zoonotic disease responsible for bacterial infections [28-29]. A strict quarantine is in force in other countries such as Germany, Australia, and the United States to prevent the entry of diseases from abroad which is also a crucial problem for Republic of Korea.
As the incidence rate of zoonotic bacteria in internationally transported snakes is high, it is suggested that in the trading of wild animals, including snakes, inspections should be more firmly conducted before and after transportation according to strict quarantine guidelines similar to those in advanced countries. Additionally, based on the study of antibiotic resistance, we should prepare against a zoonotic outbreak as the result of transported snakes. By accurately characterizing the microflora in reptiles that have the potential to induce zoonosis, they can continue to be safely provided as experimental laboratory animals and zoonotic outbreaks in the public as well as veterinarians can be avoided.
Acknowledgments
This work was supported by a grant (code: 20070401034011) from BioGreen 21 Program, Rural Development Administration, Republic of Korea.
References
1. Weissman T, Noctor SC, Clinton BK, Honig LS, Kriegstein AR. Neurogenic radial glial cells in reptile, rodent and human: from mitosis to migration. Cerebral Cortex. 2003; 13(6):550–559. PMID: 12764028.

2. Chapouton P, Jagasia R, Bally-Cuif L. Adult neurogenesis is non-mammalian vertebrates. Bioessays. 2007; 29:745–757. PMID: 17621643.
3. Handrigan GR, Leung KJ, Richman JM. Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development. 2010; 137:3545–3549. PMID: 20876646.

4. Naumann GB, Silva LF, Silva L, Faria G, Richardson M, Evangelista K, Kohlhoff M, Gontijo CM, Navdaev A, de Rezende FF, Eble JA, Sanchez EF. Cytotoxicity and Inhibition of platelet aggregation caused by an 1-amino acid oxidase from Bothrops leucurus venom. Biochim Biophys Act. 2011; 1810(7):683–694.
5. Zhou H, Dou J, Wang J, Chen L, Wang H, Zhou W, Li Y, Zhou C. The antibacterial activity of BF-30 in vitro and in infected burned rats is through interference with cytoplasmic membrane integrity. Peptides. 2011; 32(6):1131–1138. PMID: 21515321.
6. Baron S. Iglewsk BH, editor. Pseudomonas. Medical Microbiology. 1996. 4th ed. Galveston: University of Texas Medical Branch at Galveston;Chapter 27 (http://www.ncbi.nlm.nih.gov/books/NBK7627).
7. Blaylock RS. Normal oral bacterial flora from some southern African snakes. Onderstepoort J Vet Res. 2001; 68(3):175–182. PMID: 11769348.
8. Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010; 23(1):35–73. PMID: 20065325.
9. Gilmore MS, Ferretti JJ. Microbiology. The thin line between gut commensal and pathogen. Science. 2003; 299(5615):1999–2002. PMID: 12663906.
10. Endimiani A, Luzzaro F, Brigante G, Perilli M, Lombardi G, Amicosante G, Rossolini GM, Toniolo A. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2005; 49:2598–2605. PMID: 15980325.
11. Sattler CA, Mason EO Jr, Kaplan SL. Nonrespiratory Stenotrophomonas maltophilia infection at a children's hospital. Clin Infect Dis. 2000; 31:1321–1330. PMID: 11095997.
12. Koneman EW, Allen SD, Janda WM, Schrekenberger PC, Winn WC. Color Atlas and Textbook of Diagnostic Microbiology. 1997. 5th ed. Philadelphia: Lippincott Williams & Wilkins;p. 1395.
13. Hendry DI. Clinical Microbiology Procedure Handbook. 2003. 2nd ed. Washington DC: ASM Press;p. 19–55.
14. Svejgaard E. The role of microorganisms in atopic dermatitis. Semin Dermatol. 1990; 9(4):255–261. PMID: 2285570.
16. Nishioka Sde A, Silveira PV. Bacteriology of abscesses complicating bites of lance-headed vipers. Ann Trop Med Parasitol. 1992; 86:89–91. PMID: 1616400.
17. Liao WB, Lee CW, Tsai YS, Liu BM, Chung KJ. Influential factors affecting prognosis of snakebite patients' management: Kaohsiung Chang Gung Memorial Hospital experience. Chang Gung Med J. 2000; 23:577–583. PMID: 11126148.
18. Mahajan RK, Khan SA, Chandel DS, Kumar N, Hans C, Chaudhry R. Fatal case of Salmonella enteric subsp. arizonae gastroenteritis in an infant with microcephaly. J Clin Microbiol. 2003; 41(12):5830–5832. PMID: 14662995.
19. Stepanovic S, Jezek P, Vukovic D, Dakic I, Petras P. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J Clin Microbiol. 2003; 41(11):5262–5264. PMID: 14605178.
20. Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of enterococcus: An emerging nosocomial pathogen. Indian J Med Microbiol. 2009; 27(4):301–305. PMID: 19736397.
21. Shek KC, Tsui KL, Lam KK, Crow P, Ng KH, Ades G, Yip KT, Grioni A, Tan KS, Lung DC, Lam TS, Fung HT, Que TL, Kam CW. Oral bacterial flora of the Chinese cobra (Naja atra) and bamboo pit viper (Trimeresurus albolabris) in Hong Kong SAR, China. Hong Kong Med J. 2009; 15(3):183–190. PMID: 19494373.
22. Voelz A, Muller A, Gillen J, Le C, Dresbach T, Engelhart S, Exner M, Bates CJ, Simon A. Outbreaks of Serratia marcescens in neonatal and pediatric intensive care units: clinical aspects, risk factors and management. Int J Hyg Environ Health. 2010; 213(2):79–87. PMID: 19783209.
24. Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005; 7(11):1673–1685. PMID: 16232283.

25. Huet LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, Li X, Li JB. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents. 2011; 37:230–234. PMID: 21296557.
26. Goss CH, Otto K, Aitken ML, Rubenfeld GD. Detecting Stenotrophomonas maltophilia does not reduce survival of patients with cystic fibrosis. Am J Respir Crit Care Med. 2002; 166:356–361. PMID: 12153970.
27. Garg A, Sujatha S, Garg J, Acharya NS, Chandra Parija S. Wound infections secondary to snakebite. J Infect Dev Ctries. 2009; 3(3):221–223. PMID: 19759478.

28. Tu ZC, Zeitlin G, Gagner JP, Keo T, Hanna BA, Blaser MJ. Campylobacter fetus of reptile origin as a human pathogen. J Clin Microbiol. 2004; 42(9):4405–4407. PMID: 15365057.
29. Wang F, Zhou L, Gong S, Deng Y, Zou J, Wu J, Liu W, Hou F. Severe infection of wild-caught snakes with Spirometra erinaceieuropaei from food markets in Guangzhou. China involves a risk for zoonotic sparganosis. J Parasitol. 2011; 97(1):170–171. PMID: 21348631.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download