Abstract
Gomisin A possesses a hepatic function-facilitating property in liver-injured rats. Its preventive action on carbon tetrachloride-induced cholestasis is due to maintenance of the function of the bile acids-independent fraction. To investigate alterations in gene expression after gomisin A treatment on injured rat liver, DNA microarray analyses were performed on a Rat 44K 4-Plex Gene Expression platform with duplicated reactions after gomisin A treatment. We identified 255 up-regulated and 230 down-regulated genes due to the effects of gomisin A on recovery of carbon tetrachloride-induced rat liver damage. For functional characterization of these genes, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes biochemical pathways analyses were performed. Many up-regulated or down-regulated genes were related to cell cycle or focal adhesion and cell death genes, respectively. Our microarray experiment indicated that the liver repair mechanism induced by gomisin A was strongly associated with increased gene expressions related to cell cycle and suppression of the gene expression related in cell death.
Schisandra chinensis (Turcz.) Baill grows wild in the easternmost parts of Russia, the Kuril islands, Southern Sachalin, Northeastern China, Korea and Japan [1]. S. chinensis has some beneficial effects, including hepatoprotective, antioxidative, anti-inflammatory, anticancer, and anti-HIV actions. S. chinensis has many active ligands, including gomisins A, B, C, F, G, H, J, M and N, and schisandrin B and C [2]. These ligands isolated from S. chinensis showed protective effects from inflammatory infiltration and liver cell necrosis induced by carbon tetrachloride (CCl4) [3], including acetaminophen-induced hepatotoxicity and glutamate-induced oxidative neuronal damage [4,5]. In particular, gomisin A has been known to possess a liver function-facilitating property that is related to preventive action on CCl4-induced fibrosis in liver-injured rats by maintaining the function of the bile acids-independent fraction [6]. In addition, gomisin A has been reported to be effective in improving immunologically induced acute hepatic failure [7], as well as in stimulating liver regeneration after a partial hepatectomy [8].
Liver fibrosis is characterized by excessive deposition of connective tissue and distortion. Many mediators are involved in the process of fibrogenesis. Molecular mechanisms involved in fibrogenesis revealed that transforming growth factor-β (TGF-β) played a pivotal role, and depletion of fibrosis by regulating the expression of TGF genes is expected to be a new therapy for liver fibrosis. For example, taurine, heparin-superoxide dismutase conjugate, tetramethyl pyrazine, imatinib mesylate, perindopril, and ginkgo biloba down-regulated the TGF pathway [9-14]. These therapy molecules protect the rat liver from fibrogenesis induced by CCl4, and the possible mechanism could involve the down-regulation of TGF-β.
As the human genome project has been completed, attention is currently focused on understanding the gene expression profiles of disease states in cells and tissues, as well as the development of platform technology or methodology for detecting and quantitating gene expression levels. Northern blots, Southern blots, PCR, S1 nuclease protection, differential display, cDNA library sequencing, and serial analysis of gene expression (SAGE) methods have limited ability to analyze a large amount of data quantitatively. The DNA microarray system is one of the most powerful technologies for analyzing gene expression in many fields of biological research analyzing the expression profiles of thousands of genes in a wide range of biological systems [15-19]. This technology enables scientists to do a high-sensitivity parallel screening of a large number of genes with a small amount of starting material. Recently, the introduction of fluorescent probes has made it possible to array tens of thousands of short oligo-nucleotides representing the full transcriptome of a species on a miniaturized slide-glass array [20].
In the present work, we investigated the transcriptome profile related to the hepatoprotective effects of gomisin A on CCL4-induced rat liver damage. Using microarray technology, we screened for genes differentially expressed after treatment of gomisin A on rat livers that were damaged by CCl4. DNA microarray-based gene profiling identified 255 up-regulated genes and 230 down-regulated genes, and their specific metabolic pathways were described.
The fruits of S. chinensis used in this study were collected from Moongyeong, Korea in September, 2005. A voucher specimen (accession No. SC-PNUNPRL-1) was deposited in the Herbarium of Pusan National University. Pure gomisin A was identified by high performance liquid chromatography on a Phenomenex Luna C18 column (150×4.6 mm internal diameter, 5-µm particle size; Phenomenex, Torrance, CA, USA) [21]. The chemical structure of gomisin A used in this study was verified by liquid chromatography-mass spectrometry (LC-MS; Bruker BioApex FT Mass Spectrometer, Billerica, MA, USA) and nuclear magnetic resonance (NMR) analysis (Varian Inova 500 Spectrometer, Vernon Hills, IL, USA). Optical rotations were recorded on a Jasco DIP-370 Digital polarimeter, Essex, UK). IR spectra were recorded on an ATI Mattson Genesis Series FTIR, Golden Valley, MN, USA). NMR spectra (1H, 13C) were recorded in CDCl3 on a Varian Inova 500 Spectrometer operating at 500 MHz for 1H and 125 MHz for 13C, running gradients and using residual solvent peaks as internal references. High-resolution mass spectra were recorded on a Bruker BioApex FT Mass Spectrometer.
All subjects were female Sprague-Dawley (SD) rats, purchased from Samtaco Biokorea (Osan, Korea). These rats were kept in Pusan National University/Laboratory Animal Resources Center in accordance with the US National Institute of Health (NIH) Animal Care policies. The rats were given a standard irradiated chow diet (Purina Mills, St. Louis, MO, USA) ad libitum and were maintained in a specified pathogen-free state under a strict light cycle (light on at 06:00 and off at 18:00). Furthermore, the animal protocol used in this study has been reviewed and approved by the Pusan National University Institutional Animal Care and Use Committee (PNU-IACUC).
Eight-week-old SD rats were randomly divided into four groups with five rats per group. The first group of SD rats was not treated by any compounds as an untreated control group (UT group), while the second group received 100 mg/kg body weight per day of gomisin A via oral administration for 4 days (gomisin A-treated group, GT group). The third group received a comparable volume of olive oil via oral gavage daily, while the fourth group received 100 mg/kg body weight per day of gomisin A via oral administration for 4 days. At the fifth day, all animals in the first and second groups were immediately euthanized using CO2 gas and the liver samples collected. Meanwhile, the third (CCl4-treated group, CT group) and fourth groups (gomisin A and CCl4-treated group, GCT group) received 0.1 mL of CCl4 (Sigma-Aldrich, St. Louis, MO, USA) solution via intraperitoneal injection. After 24 hours post-CCl4 injection, these animals were immediately euthanized using CO2 gas. Liver samples were collected and stored in Eppendorf tubes at -70℃ until assayed.
Liver tissues collected from rats were fixed with 10% formalin for 12 hours, embedded in paraffin wax, and then sectioned into 5 µm-thick slices. The liver sections were stained with hematoxylin and eosin (H&E, Sigma-Aldrich) and morphological features were observed under light microscopy (Leica Mycrosystems, Heerbrugg, Switzerland).
Total liver RNA was isolated from rat liver tissue using Trizol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions. The total RNA was dissolved in diethyl pyrocarbonate-treated distilled water, and stored at -70℃. DNA microarray analyses were performed according to the manufacturer's standard protocol. Briefly, Cy5-dUTP-labeled cRNA were prepared from 200 µg of total RNA isolated from a liver of each group. Complementary RNA was labeled using a fluorescent cRNA labeling kit (Agilent, Delaware, DE, USA) according to the manufacturer's instructions. Then, 10X blocking agent and ×25 fragmentation buffer were added to the labeled cRNAs by following the instructions from the hybridization kit (Agilent). For hybridization, total cRNAs were applied to a microarray (Rat 44K 4-Plex Gene Expression platform; Agilent) followed by incubation at 65℃ overnight under humidified conditions. To control the labeling differences, technical replication was conducted in which duplicated reactions were carried out by switching over the fluorescent dyes. Microarray analysis was performed on using total cRNA hybridization images captured by a microarray scanner system (G2545CA; Agilent) using feature extraction (FE) 9.5.3 software at the scanning resolution of 5 mm.
Analysis of feature extraction text files produced by the Agilent scanner and further data evaluations were performed by using the Genespring GX7.3.1 software package (Agilent Technologies, Santa Clara, CA, USA). Relative expression levels of genes were calculated after normalization of the signal intensity values and outputted with the unigene and genebank descriptors to a Microsoft Excel data spreadsheet. Gene ontology (GO) analysis based on biological process and molecular function ontology analysis was performed using the web tool Database for Annotation, Visualization, and Integrated Discovery (DAVID). Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathways analysis was also performed by the web tool KEGG (http://www.genome.jp/kegg/pathway.html).
To investigate the protective effects of gomisin A on CCl4-induced hepatocellular damage, we examined the histopathology of livers from all four groups. In the GCT group rat liver, histological alteration of liver tissue was observed. In both the UT group (Figure 1A) and GT group (Figure 1B), the histopathology showed a normal distribution of hepatocytes with clear visible nuclei, portal triads, and central veins. After CCl4 treatment, extensive centrolobular necrosis was observed in and around the central vein of liver (Figure 1C). However, necrosis was significantly decreased in the liver section of the group pretreated with gomisin A for 4 days (Figure 1D). These results suggest that pretreatment with gomisin A may protect against hepatocellular damage induced by CCl4 treatment.
Preparation of total RNA with high purity is essential for a successful microarray experiment. The quality of RNA was measured by Agilent 2100 Bioanalyzer (Agilent). The Agilent 2100 Bioanalyzer provides a platform for the electrophoresis of nucleic acids on a disposable chip [22,23]. Each RNA sample (1 mL) was loaded into individual wells on a chip and electrophoresed in the Bioanalyzer. 18s and 28s ribosomal peaks were present in all samples by giving RNA Integrity Numbers (RINs) from 8.0 to 10.0, which indicated the verification of a good quality of RNA samples for microarray analysis.
Liver RNA samples from three individual rats were pooled for each experimental group. Each RNA sample was hybridized with 41,012 probes on Rat 44K 4-Plex Gene Expression platform. Systematic errors across all variables in an experiment can be attributed to well-known sources including biases linked to the different dyes used at the labeling step. Possible systemic bias in the data was removed by signal dependent LOWESS normalization of the MA-plot [24]. This normalized data were tested for significant gene expression fold changes between control and samples by a t-test.
We identified a number of differentially expressed genes in the GCT group. In total, the expression of 2,305 genes was up-regulated more than two-fold (>2), while the expression of 875 genes was down-regulated more than two-fold (<0.5). To narrow down the number of genes that are possibly involved in gomisin A-induced recovery from CCl4-induced injury, a selection strategy (Figure 2) was employed, and finally, 255 up-regulated and 230 down-regulated genes were selected. The up-regulated 255 genes were the genes that showed increased expression level (>2) after the treatments in the GCT group, but showd a decreased expression level in both the GT group (<2) and CT group (<1). The down-regulated 230 genes were the genes for which expression level were decreased (<0.5) in GCT group but were not significantly decreased in either the GT group (>0.5) or the CT group (>1) compared to the UT group (Figure 2).
Hierarchical cluster analysis of 488 selected genes resulted in Clusters I and II, which comprise 255 and 230 genes that were up- or down-regulated, respectively, by the treatment of gomisin A on the rat liver damaged by CCl4. A closer relation was observed between the GT group and GCT group, indicating that many genes influenced by gomisin A treatment on the normal liver may be associated with recovery from the liver damage induced by CCl4 (Figure 3).
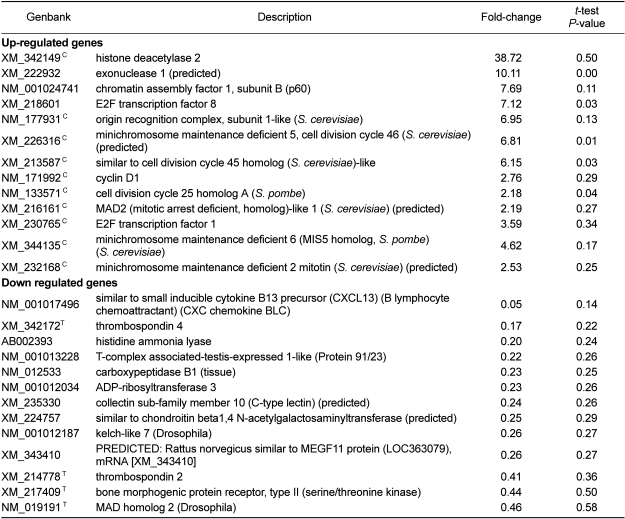
Among 488 up- or down-regulated genes, 26 genes that were high ranked in the fold change are listed in Table 1. A majority of up-regulated genes were related to cell cycle. These genes included histone deacetylase, which removes acetyl groups and leads to the compaction of chromatin and DNA silencing. Histone deacetylase has been shown to be recruited by the retinoblastoma protein (Rb) and related pocket proteins to the promoters of several cell cycle genes. Thus, the histone deacetylase gene has been implicated in controlling the transcription of core cell cycle regulators [25,26]. Up-regulated genes involved chromatin assembly factor (CAF) as well. CAF is essential for normal cell cycle maintenance. CAF1 loss of function in mammalian cells leads to the activation of a DNA-damage signaling pathway that slows down the S-phase of cell division and arrests the cell cycle [27]. E2F is also involved in cell cycle maintenance [28]. Most of the down-regulated genes were related to focal adhesions and cell death. These genes included thrombospondin (TSP), which is a member of a group of extracellular matrix (ECM) proteins. When exposed to cells in its soluble form, thrombospondin has primarily anti-adhesive effects characterized by a reorganization of stress fibers and loss of focal adhesion plaques as ascertained by interference reflection microscopy [29-31]. Down-regulated genes also included ADP ribosyltransferase (ADPRT). ADPRT activity interferes with eukaryotic DNA synthesis and endocytosis and causes cytotoxicity and cell death of mammalian cells [32,33].
Gene ontology and biological pathway analysis were performed with the genes selected as being specifically regulated in the process of gomisin A-induced recovery of rat liver tissues. Among 255 up-regulated and 230 down-regulated genes, 110 and 141 genes with Entrez gene IDs were matched to genes in Rattus norvegicus, respectively. These genes were further analyzed for functional classification by GO term and pathway assignment by KEGG.
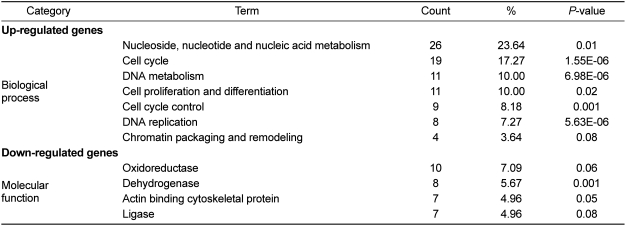
The most statistically significantly represented GO terms are displayed in Table 2. "Nucleic acid metabolism and cell cycle" was identified as the most significantly related biological process to the up-regulated genes. For down-regulated genes, "oxidoreductase and dehydrogenase" were identified as the most significantly related molecular functions. Oxidoreductases belong to a family of genes that are involved in the regulation of programmed cell death. Oxidoreductase mediates cell death caused by the novel anti-cancer drug β-lapachone [34]. Also, dehydrogenase causes a programmed cell death in ERK kinase signaling events [35].
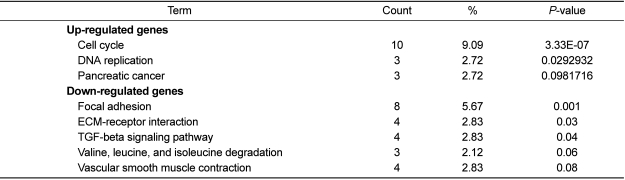
Biological pathway assignment based on KEGG was displayed in Table 3. "Cell cycle" (Figure 4A) was identified by up-regulated genes as the most statistically significant pathway, which was in accordance with the result of GO term analysis. While the majority of up-regulated genes were related to cell cycle in the distribution of regulated genes, focal adhesion, ECM-receptor interaction, and TGF-beta signaling pathway (Figure 4B) were identified as the most statistically significant in down-regulated genes. A single CCl4 administration can cause hepatic injury that is associated with ECM accumulation. During the remodeling process of the liver architecture from hepatic damage, including inflammation, fibrosis and regeneration, TGF-β1 plays a pivotal role in stimulating the liver [36]. Initiation of hepatic fibrosis starts with paracrine stimulation of injured hepatocytes and endothelial cells, as well as subtle changes in ECM composition [37]. They produce pro-fibrogenic cytokines such as TGF-β. TGF-β as a major fibrogenic cytokine is heavily involved in abnormal ECM accumulation by increasing the synthesis of ECM components [38]. ECM molecules bind to integrin cell surface receptors and activate downstream focal adhesion molecules involved in the regulation of anchorage-dependent cell growth, proliferation, survival, differentiation, morphology, migration, and death [39,40]. Especially, liver fibrosis is characterized by excessive deposition of ECM, which leads to a severe pathological disturbance in the liver. During liver fibrogenesis, an increase in TGF-beta activity has been reported [41]. TGF-beta is an important cytokine in regulating the production of ECM for liver fibrosis [42,43].
In conclusion, our histological observation showed that gomisin A treatment could repair the rat liver with CCL4-induced damage. Further, our microarray experiment indicated that the liver repair mechanism induced by gomisin A was strongly associated with increased expression of genes related to cell cycle and suppression of the genes involved in cell death mediated by the TGF-β pathway.
References
1. Liu GT. Biochemical and pharmacologic effects of wuweizi [Schizandra chinensis (Turcz.) Baill.] and its chemical contents on animal livers. Sheng Li Ke Xue Jin Zhan. 1988; 19(3):197–203. PMID: 3074486.
2. Suprunov NI, Vetlugina IV. Determination and study of lignan distribution in the fruits of Schisandra chinensis (Turcz.) Baill. Farmatsiia. 1972; 21(3):34–37. PMID: 5035640.
3. Bao TT, Liu GT, Song ZY, Xu GF, Sun RH. A comparison of the pharmacologic actions of 7 constituents isolated from Fructus schizandrae. Chin Med J. 1980; 93(1):41–47. PMID: 6768500.
4. Maeda S, Takeda S, Miyamoto Y, Aburada M, Harada M. Effects of gomisin A on liver functions in hepatotoxic chemicals-related rats. Jpn J Pharmacol. 1985; 38(4):347–353. PMID: 4068375.
5. Yamada S, Murawaki Y, Kawasaki H. Preventive effect of gomisin A, a lignan component of shizandra fruits, on acetaminophen-induced hepatotoxicity in rats. Biochem Pharmacol. 1993; 46(6):1081–1085. PMID: 8216352.

6. Maeda S, Takeda S, Miyamoto Y, Aburada M, Harada M. Effects of Gomisin A on liver functions in hepatotoxic chemicals-treated rats. Jpn J Pharmacol. 1985; 38(4):347–353. PMID: 4068375.

7. Mizoguchi Y, Shin T, Kobayashi K, Morisawa S. Effect of gomisin A in an immunologically-induced acute hepatic failure model. Planta Med. 1991; 57(1):11–14. PMID: 2062950.

8. Kubo S, Ohkura Y, Mizoguchi Y, Matsui-Yuasa I, Otani S, Morisawa S, Kinoshita H, Takeda S, Aburada M, Hosoya E. Effect of Gomisin A (TJN-101) on liver regeneration. Planta Med. 1992; 58(6):489–492. PMID: 1484885.

9. Yoshiji H, Kuriyama S, Noguchi R, Ikenaka Y, Yoshii J, Yanase K, Namisaki T, Kitade M, Yamazaki M, Asada K, Akahane T, Tsujimoto T, Uemura M, Fukui H. Amelioration of liver fibrogenesis by dual inhibition of PDGF and TGF-β with a combination of imatinib mesylate and ACE inhibitor in rats. Int J Mol Med. 2006; 17(5):899–904. PMID: 16596278.

10. Liu SQ, Yu JP, Chen HL, Luo HS, Chen SM, Yu HG. Therapeutic effects and molecular mechanisms of Ginkgo biloba extract on liver fibrosis in rats. Am J Chin Med. 2006; 34(1):99–114. PMID: 16437743.
11. Ding J, Yu J, Wang C, Hu W, Li D, Luo Y, Luo H, Yu H. Ginkgo biloba extract alleviates liver fibrosis induced by CCl4 in rats. Liver Int. 2005; 25(6):1224–1232. PMID: 16343076.
12. Liu J, Tan H, Sun Y, Zhou S, Cao J, Wang F. The preventive effects of heparin-superoxide dismutase on carbon tetrachloride-induced acute liver failure and hepatic fibrosis in mice. Mol Cell Biochem. 2009; 327(1-2):219–228. PMID: 19242656.

13. Liang J, Zhang XL, Yang GY, Pang YS, Yuan HF, Liang JS, Huang RB. Observation of the promotion effect taurine on hepatic stellate cell's apoptosis in rat hepatic fibrosis model. Sichuan Da Xue Xue Bao Yi Xue Ban. 2005; 36(3):365–367. PMID: 15931870.
14. Lu B, Yu L, Li S, Si S, Zeng Y. Alleviation of CCl4-induced cirrhosis in rats by tetramethylpyrazine is associated with down regulation of leptin and TGF-β1 pathway. Drug Chem Toxicol. 2010; 33(3):310–315. PMID: 20433334.
15. Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996; 6(7):639–645. PMID: 8796352.

16. Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996; 93(20):10614–10619. PMID: 8855227.

17. Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA. 1999; 96(23):13264–13269. PMID: 10557309.

18. Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999; 21(1):10–14. PMID: 9915494.

19. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999; 21(1):33–37. PMID: 9915498.

20. Jordan BR. Large-scale expression measurement by hybridization method: from high-density membranes to 'DNA chips'. J Biochem. 1998; 124(2):251–258. PMID: 9685711.
21. Avula B, Dentali S, Khan IA. Simultaneous identification and quantification by liquid chromatography of benzethonium chloride, methyl paraben and triclosan in commercial products labeled as grapefruit seed extract. Pharmazie. 2007; 62(8):593–596. PMID: 17867553.
22. Panaro NJ, Yuen PK, Sakazume T, Fortina P, Kricka LJ, Wilding P. Evaluation of DNA fragment sizing and quantification by the agilent 2100 bioanalyzer. Clin Chem. 2000; 46(11):1851–1853. PMID: 11067828.

23. Funes-Huacca M, Regitano LC, Mueller O, Carrilho E. Semiquantitative determination of Alicyclobacillus acidoterrestris in orange juice by reverse-transcriptase polymerase chain reaction and capillary electrophoresis-laser induced fluorescence using microchip technology. Electrophoresis. 2004; 25(21-22):3860–3864. PMID: 15565670.
24. Yang HY, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acid Res. 2002; 30(4):e15. PMID: 11842121.

25. Goodsell DS. The molecular perspective: Histone deacetylase. Oncologist. 2003; 8(4):389–391. PMID: 12897336.

26. Stadler JA, Shkumatava A, Norton WH, Rau MJ, Geisler R, Fischer S, Neumann CJ. Histone deacetylase 1 is required for cell cycle exit and differentiation in the Zebrafish retina. Dev Dyn. 2005; 233(3):883–889. PMID: 15895391.

27. Haushalter KA, Kadonaga JT. Chromatin assembly by DNA translocating motors. Nat Rev Mol Cell Biol. 2003; 4(8):613–620. PMID: 12923523.
28. Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys. 2003; 412(2):157–169. PMID: 12667479.

29. Greenwood JA, Murphy-Ullrich JE. Signaling of de-adhesion in cellular regulation and motility. Microsc Res Tech. 1998; 43(5):420–432. PMID: 9858339.

30. Murphy-Ullrich JE, Höök M. Thrombospondin modulates focal adhesions in endothelial cells. J Cell Biol. 1989; 109(3):1309–1319. PMID: 2768342.

31. Murphy-Ullrich JE, Lane TF, Pallero MA, Sage EH. SPARC mediates focal adhesion disassembly in endothelial cells through a follistatin-like region and the Ca2+-binding EF-hand. J Cell Biochem. 1995; 57(2):341–350. PMID: 7539008.
32. Barbieri AM, Sha Q, Bette-Bobillo P, Stahl PD, Vidal M. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect Immun. 2001; 69(9):5329–5334. PMID: 11500402.
33. Pederson KJ, Barbieri JT. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol Microbiol. 1998; 30(4):751–759. PMID: 10094623.
34. Dong GZ, Youn H, Park MT, Oh ET, Park KH, Song CW, Choi EK, Park HJ. Heat shock increases expression of NAD(P)H:quinone oxidoreductase (NQO1), mediator of β-lapachone cytotoxicity, by increasing NQO1 gene activity and via Hsp70-mediated stabilisation of NQO1 protein. Int J Hyperthermia. 2009; 25(6):477–487. PMID: 19657853.

35. Fico A, Paglialunga F, Cigliano L, Abrescia P, Verde P, Martini G, Iaccarino I, Filosa S. Glucose-6-phosphate dehydrogenase plays a crucial role in protection from redox-stress-induced apoptosis. Cell Death Differ. 2004; 11(8):823–831. PMID: 15044966.

36. Kim SJ, Park JH, Kim KH, Lee WR, Chang YC, Park KK, Lee KG, Han SM, Yeo JH, Pak SC. Bee venom inhibits hepatic fibrosis through suppression of pro-fibrogenic cytokine expression. Am J Chin Med. 2010; 38(5):921–935. PMID: 20821823.

37. Gressner AM, Bachem MG. Molecular mechanisms of liver fibrogenesis--a homage to the role of activated fat-storing cells. Digestion. 1995; 56(5):335–346. PMID: 8549875.
38. Tahashi Y, Matsuzaki K, Date M, Yoshida K, Furukawa F, Sugano Y, Matsushita M, Himeno Y, Inagaki Y, Inoue K. Differential regulation of TGF-β signal in hepatic stellate cells between acute and chronic rat liver injury. Hepatology. 2002; 35:49–61. PMID: 11786959.

39. Howe A, Aplin AE, Alahari SK, Juliano RL. Integrin signaling and cell growth control. Curr Opin Cell Biol. 1998; 10(2):220–231. PMID: 9561846.

40. Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999; 285(5430):1028–1032. PMID: 10446041.

41. Bataller R, Gäbele E, Parsons CJ, Morris T, Yang L, Schoonhoven R, Brenner DA, Rippe RA. Systemic infusion of angiotensin II exacerbates liver fibrosis in bile duct-ligated rats. Hepatology. 2005; 41(5):1046–1055. PMID: 15841463.

42. Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000; 275(4):2247–2250. PMID: 10644669.

43. De Bleser PJ, Xu G, Rombouts K, Rogiers V, Geerts A. Glutathione levels discriminate between oxidative stress and transforming growth factor-β signaling in activated rat hepatic stellate cells. J Biol Chem. 1999; 274(48):33881–33887. PMID: 10567349.

Figure 1
Histology of liver tissue. A, vehicle; B, gomisin A alone; C, carbon tetrachloride (CCl4) alone; D, CCl4+gomisin A. The hepatocytic morphology on the liver section stained with hematoxylin and eosin was observed with light microscope at ×400. Arrows indicate the necrotic hepatocytes.

Figure 2
Dendrograms showing strategies for identifying specifically regulated genes in the process of gomisin A-induced recovery of rat liver tissues; 255 up-regulated (A) and 230 down-regulated (B) genes were identified. In A, <1 (in CT) and <2 (in GT) indicate the number of genes that did not showed significant up-regulation in CT and GT groups, respectively, and >2 (in GCT) indicates the number of genes that showed significant up-regulation in GCT group. In B, >1 (in CT) and >0.5 (in GT) indicate the number of genes that did not showed significant down-regulation in CT and GT treated groups, respectively, and >0.5 (in GCT) indicates the number of genes that showed significant down-regulation in GCT group.

Figure 3
Hierarchical clustering of 488 genes selected as shown in Figure 1. Y-axis shows the list of genes and X-axis shows control and samples. Red fields show un-regulation of genes (absolute difference) and green fields show down-regulation of genes. U, UT group; C, CT group; G, GT group; GC, GCT group.

Figure 4
Pathway assignment of the important genes based on KEGG. The genes of up-regulation in cell cycle pathway (A) and the genes of down-regulation in TGF-β signaling pathway (B) were determined by KEGG pathway. The genes found in our microarray analysis were labeled by star symbols.

Table 1
The list of genes identified by the analysis shown in Figure 2. Only the 26 genes that were high ranked in the fold change and/or assigned in the pathway based on KEGG are shown (Figure 4). The genes involved in the cell cycle pathway or TGF-β signaling pathway were indicated by C, or T, respectively




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download