Abstract
Mycoplasmas are highly fastidious bacteria, difficult to culture and slow growing. Many species of mycoplasmas are important pathogens that cause respiratory infection in laboratory animals and that are known to affect experimental results obtained with contaminated animals. The aim of the present study was to develop a sensitive and specific assay for the detection of mycoplasma species. To this end, we developed a polymerase chain reaction and dot blot hybridization assay (PCR/DBH) for detecting mycoplasma DNA and evaluated it for its sensitivity and specificity. Mycoplasma consensus primer pairs were used for the amplification of target DNA. When PCR product was visually detected, the limit of detection of the PCR test was 102 pg of mycoplasma purified DNA. For DBH, the amplified DNA was labeled by incorporation of digoxigenin (DIG). This DIG-labeled probe was capable of detecting 104 pg of purified mycoplasma DNA by DBH. PCR/DBH was more sensitive than PCR or DBH alone and was also very specific. Our PCR/DBH assay can be applied efficiently to confirm the presence of mycoplasma species on clinical samples and to differentiate between mycoplasma species infection and other bacterial infections.
Mycoplasmas belong to the class Mollicutes and are among the smallest free-living microorganisms capable of auto-replication. Mycoplasmas are highly fastidious bacteria, difficult to culture and slow growing [1]. Many species are important veterinary pathogens causing respiratory infection, mastitis, conjunctivitis, arthritis, and occasionally abortion [1]. Also, in the field of laboratory animal medicine, it has been reported that mycoplasma infections are very common and considered highly contagious [2]. Major mycoplasmas of laboratory animals include Mycoplasma (M.) hyopneumoniae, M. pulmonis, M. collies, M. neurolyticum, M. arthritiditis, M. hyorhinis, and M. mycoides [3]. Identification of mycoplasmas as the causative agents of disease is often hindered by the lack of a rapid diagnostic test together with similarities in the clinical diseases that they cause. Conventional methods of diagnosis are based on culture and serological tests, such as the complement fixation test [4], enzyme-linked immunosorbent assays [5], and immunoblotting [6], and can be time-consuming, insensitive, and nonspecific. It is necessary to clarify the current status of mycoplasma contamination in laboratory animal colonies because mycoplasmas are prevalent in animals in commercial and research facilities [7].
Mycoplsama infection in laboratory animals interferes with biomedical research [8]. The transmission of M. pulmonis by aerosol from an infected to a healthy rat by sneezing at a distance of about 120 cm strongly suggests that such transfers may happen between rats and humans [2]. The technicians cleaning cages of facilities with rats infected with M. pulmonis are liable to be infected with these bacteria. Although mycoplasmas show specificity for their hosts, isolation of some mycoplasmas from unusual hosts has already been reported [9,10].
Polymerase chain reaction (PCR) provides a powerful technique for identifying different mycoplasmas and studying homology between their nucleic acids. However, those kinds of PCR assays require multiple assays because there are a lot of mycoplasma species [1]. Also, PCR has the high risk of the false-positive reaction by contamination and of the false-negative result by enzymatic inhibitors [11-13]. In order to avoid problems related to nucleic acid amplification, efforts have been made to develop specific hybridization assays such as dot blot hybridization (DBH) and in situ hybridization [14]. DBH is a simple and specific method for detection of pathogens and has been reported to be a method with higher specificity and lower sensitivity compared to PCR assays [15-17].
The aims of the present study were to develop a mycoplasma genus-specific PCR/DBH assay using a 16S ribosomal DNA gene. Such an assay might be a fast and practical method for detection and identification of Mycoplsama species.
M. hyopneumoniae (ATCC 25934) was obtained from the American Type Culture Collection (ATCC, Rockville, USA). The mycoplasmas were grown in modified Friis medium [18], containing 20% porcine serum (Gibco BRL, Rockville, USA), 5% fresh yeast extract (Gibco-BRL), methicillin (0.15 mg/mL; Sigma-Aldrich Canada, Oakville, Canada), bacitracin (0.15 mg/mL; Sigma-Aldrich Canada), and thallium acetate (0.08 mg/mL; Sigma-Aldrich). The cells were harvested by centrifugation at 12,000 g for 30 min at 4℃, washed three times, and suspended in 0.1M phosphate-buffered saline (PBS), pH 7.4.
M. hyopneumoniae DNA was extracted from cultured M. hyopneumoniae using an AccuPrep Genomic DNA extraction kit (Bioneer, Daejeon, Korea) according to the manufacturer's instructions. The DNA was eluted in Tris-EDTA buffer (pH 8.0), and an aliquot was used for the PCR amplification. Amplification of the V3 region of the 16S ribosomal DNA was performed with consensus primers GC-341F (5'-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG) and 534R (5'-ATT ACC GCG GCT GCT GG), which were based on the sequences reported by Weisburg et al. [19]. The template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer) containing 2.5 U of Taq DNA polymerase, 250 µM each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and the gel loading dye. The volume was adjusted with distilled water to 20 µL. The reaction mixture was subjected to denaturation at 94℃ for 5 min followed by 30 cycles of 95℃ for 1 min, 55℃ for 45 sec, and 72℃ for 1 min, and then a final extension step of 72℃ for 10 min was done. Samples were kept at 4℃ until analysis. Reactions were conducted using a My Genie 32 Thermal Block PCR (Bioneer).
The mycoplasma consensus DNA probes were constructed by PCR and labeled with digoxigenin after the amplification reaction. The mycoplasma consensus PCR using 16S ribosomal DNA pairs, was performed as described previously [3]. After amplification, PCR products were purified using Wizard PCR preps (Promega Biotech, Medison, USA). Purified PCR products were labeled by random priming with digoxigenin-dUTP (Roche Applied Science, Mannheim, Germany) or by means of a commercial kit according to the manufacturer's instructions.
The detection of PCR products using a non-radioactive DNA probe was conducted by PCR/DBH assay to allow for sensitive and specific detection of target DNA. Dot blotting was achieved by direct application on a positively charged nylon membrane (Roche Applied Science). The PCR products after primary amplification were dotted on the nylon membrane. The membrane was immersed in 0.4M NaOH for 5 min and then in neutralizing buffer for 5 min. After rinsing in 2× saline-sodium citrate buffer (SSC), cross-linking between the applied DNA and the membranes was done using UV cross-linker (Stratagene, La Jolla, CA, USA). Hybridization solutions consisted of 5×SSC, 2% buffered blocking solution (Roche Applied Science), 0.1% N-lauroylsarcosine, and 0.02% sodium dodecyl sulfate. Digoxigenin (DIG)-labeled probe, which was denatured by boiling for 10 min and chilling in ice, was added in hybridization solution at 0.1 mg/mL before the hybridization. After pre-hybridization at 50℃ for 1 hour, the membrane was hybridized at 50℃ for 3 h and washed with 1×SSC at 60℃ for 10 min and 1× washing buffer (Roche Applied Science) each. For detection of hybridization, the membrane was incubated with anti-digoxigenin conjugated with alkaline phosphatase (Roche Applied Science) and then colorized with nitroblue tetrazolium (NBT) and 5-bromocresyl-3-indolyl-phosphate (BCIP). Thereafter, the development of a dark purple positive reaction was allowed to proceed for 10-30 min in the dark.
The sensitivity and specificity of PCR/DBH was evaluated. Purified M. hyopneumoniae DNA samples ranging from 107 to 101 pg were used for the primary target amplification. After amplification of those template DNA samples, PCR/DBH was done with those primary amplicons. To identify the sensitivity of primary PCR amplification alone, ten microliters of the amplified PCR products were analyzed by 2% agarose gel electrophoresis and stained with ethidium bromide for product visualization. Also, to identify the sensitivity of DBH alone, purified M. hyopneumoniae DNA samples ranging from 107 to 101 pg were directly applied on positively charged nylon membranes (Roche Applied Science) and then DBH was done. The specificity of PCR/DBH was evaluated using template DNA samples such as Actinobacillus pleuropneumoniae and Pasteurella multocida, DNAs that were provided by professor C. Chae at Seoul National University in Korea.
Amplification of the V3 region of the 16S ribosomal DNA with mycoplasma consensus primers GC-341F and 534R was performed for primary amplification of target DNA. In this study, Mycoplasma DNA was detected template DNA down to 102 pg (Figure 1). Because the samples were DNAs from cultured M. hyopneumoniae, control DNA was not need for the PCR reaction. The 16S rDNA gene (340 bp) was specifically amplified by PCR with the mycoplasma genus-specific primers (GC-341F and 534R). The strongest signal was shown in the PCR product of template DNA with 107 pg (Figure 1). The sensitivity of DBH alone was low (Figure 2). The detection limit for DBH was 104 pg mycoplasma DNAs. The highest intensity was observed in the signal of the sample DNA with 107 pg. As we decreased the amount of sample DNAs, the signal intensity became weak. A sample DNA with 103 pg did not generate any positive signals (Figure 2). PCR/DBH had higher sensitivity than PCR or DBH alone. The increased sensitivity of this technique was apparent even when sample DNA of 10 pg was detected as the positive signal and the intensities of each sample was higher than an equivalent sample that had undergone DBH alone (Figure 3). The specificity of PCR/DBH was confirmed using other bacterial DNAs with high homology in their sequences. No positive signals were observed in template DNA samples of Actinobacillus pleuropneumoniae and Pasteurella multocida in the PCR/DBH assay. However, PCR/DBH using Mycoplasma template DNA resulted in a strong positive signal (Figure 4).
The diagnosis of mycoplasma infection is usually done by cultivation of the organism or by immunofluorescence tests performed on frozen thin lung sections with polyclonal antibodies [20,21]. However, due to the fastidious nature of mycoplasma species, their culture and serological identification may take up to 1 month. Serological detection methods are further hampered by cross-reactions that have been reported between M. hyopneumoniae, M. hyorhinis, and M. flocculare [22,23]. With the advances made in molecular biology during the last few years, more is known about mycoplasma species genes. Hence, other methods can be used as diagnostic tools for this organism. Recently, PCR methods were used to detect mycoplasma species [3]. These methods are rapid and sensitive, especially if nested PCR is used. However, PCR assays are subject to a high risk of contamination through DNA carry-over and may result frequently in false positive reactions [24-26]. Thus it is necessary to improve PCR sensitivity and therefore we developed a PCR/DBH assay. That assay is a very sensitive and specific tool for mycoplasma species detection. PCR sensitivity and specificity can be increased by hybridization methods of replicated DNA with specific labeled probe. In this study, we used non-radioactive labels as DBH probes, which have made these techniques more attractive for diagnostic laboratories, because they avoid problems relative to the short half-life of radioactive compounds, their disposal, and personnel safety [27,28]. Different methods have been developed for DNA probe labeling. In the past, DNA probes were labeled by radioactive isotopes (P32, I125) but diagnostics required and initiated development of rapid, non-radioactive methods for DNA probe labeling. For non-radioactive DNA probe labeling, different enzyme and immunological markers have been used [29] such as horseradish peroxidase (HRP), monoclonal and polyclonal antibodies to the DNA probe conjugated with alkaline phosphatase, and biotin conjugated DNA probes which, in with avidin-enzymatic complex produces a yellow color that can be measured by calorimetric or densitometric methods [30]. Recently, methods have been developed for non-radioactive labeling of DNA fragments by DIG-labeled dUTP randomly incorporated in the DNA. DNA probe built-in DIG-dUTP is detected by antidigoxigenin antibodies conjugated with alkaline phosphatase, which in reaction with X-phosphate and NBT salts produces a blue-stained line where the searched hybrid is positioned in the nitrocellulose membrane. DNA probe labeling was used in this study and the method was shown to be rapid, sensitive and specific, making it suitable for the detection of primary amplified Mycoplasma species DNA products. This allowed for increased sensitivity and specificity and quantification of mycoplasma species by DNA densitometry. The total assay time including the PCR procedure and DBH detection is 8 hours. The PCR/DBH assay that was established in this study is much more sensitive and specific compared with a one step PCR assay and DFA detection.
In conclusion, we developed a sensitive and reproducible method-mycoplasma consensus PCR/DBH- for the detection of mycoplasma species DNA. The PCR/DBH assay may be a valid tool for the diagnostic laboratory and can be an alternative to PCR assays for screening of a large number of samples to detect mycoplasma species.
Acknowledgments
This research was supported by the Technology Development Program for Agriculture and Forestry (grant No. 610004-3), the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
References
1. McAuliffe L, Ellis RJ, Ayling RD, Nicholas RAJ. Differentiation of Mycoplasma species by 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis fingerprinting. J Clin Microbiol. 2003; 41(10):4844–4847. PMID: 14532239.
2. Delgado MO, Timenetsky J. Immunoblot profiles of sera from laboratory rats naturally infected with Mycoplasma pulmonis and technicians exposed to infected animal facilities. Braz J Microbiol. 2001; 32(4):301–304.

3. Kim SH, Kim O. Genus-specific polymerase chain reaction to detect mycoplasma species. Lab Anim Res. 2005; 21(3):189–192.
4. Muthomi EK, Rurangirwa FR. Passive haemagglutination and complement fixation as diagnostic tests for contagious caprine pleuropneumonia caused by the F-38 strain of mycoplasma. Res Vet Sci. 1983; 35(1):1–4. PMID: 6622834.

5. Ball HJ, Finlay D. Diagnostic application of monoclonal antibody (MAb)-based sandwich ELISAs. Methods Mol Biol. 1998; 104:127–132. PMID: 9711648.

6. Nicholas RAJ, Santini FG, Clark KM, Palmer NMA, Santis PD, Bashiruddin JB. A comparison of serological tests and gross lung pathology for the detection of contagious bovine pleuropneumonia in two groups of Italian cattle. Vet Rec. 1996; 139(4):89–93. PMID: 8843640.
7. Lindsey JR, Baker HJ, Overcash RG, Cassell GH, Hunt CE. Murine chronic respiratory disease. Significance as a research complication and experimental production with Mycoplasma pulmonis. Am J Pathol. 1971; 64(3):675–708. PMID: 4943862.
8. Davidson MK, Davis JK, Gambill GP. Whitford HW, editor. Mycoplasmas of laboratory rodents. Mycoplasmosis in Animals: Laboratory Diagnosis. 1994. Ames: Iowa State University Press;p. 97–132.
9. Armstrong CH, Yu BH, Yagoda A, Kagnoff MF. Colonization of human by Mycoplasma Canis. J Infect Dis. 1971; 124(6):607–609. PMID: 5166656.
10. Yechouron D, Lefebvre J, Robson HG, Rose DL, Tully JG. Fatal septicemia due to Mycoplasma argini: a new human zoonosis. Clin Infect Dis. 1992; 15(3):434–438. PMID: 1520790.
11. Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989; 339(6221):237–238. PMID: 2716852.

12. Biesiadecka A, Litwinska B. Application of polymerase chain reaction for the detection of herpes simplex virus DNA. Acta Microbiol Pol. 1997; 46(4):405–408. PMID: 9516988.
13. Broketa M, Vince A, Drazenovic V, Sim R, Mlinaric-Galinovic G. Non-radioactive digoxigenin DNA labeling and immunologic detection of HSV PCR products. J Clin Virol. 2001; 23(1-2):17–23. PMID: 11595580.

14. McNicol AM, Farquharson MA. In situ hybridization and its diagnostic applications in pathology. J Pathol. 1997; 182(3):250–261. PMID: 9349226.
15. Duggan MA, Inoue M, McGregor SE, Stuart GC, Morris S, Chang-Poon V, Schepansky A, Honore L. A paired comparison of dot blot hybridization and PCR amplification for HPV testing of cervical scrapes interpreted as CIN 1. Eur J Gynaecol Oncol. 1994; 15(3):178–187. PMID: 7957322.
16. Xia JQ, Yason CV, Kibenge FS. Comparison of dot blot hybridization, polymerase chain reaction, and virus isolation for detection of bovine herpesvirus-1 (BHV-1) in artificially infected bovine semen. Can J Vet Res. 1995; 59(2):102–109. PMID: 7648521.
17. Hwang SJ, Lee SD, Lu RH, Chan CY, Lai L, Co RL, Tong MJ. Comparison of three different hybridization assays in the quantitative measurement of serum hepatitis B virus DNA. J Virol Methods. 1996; 62(2):123–129. PMID: 9002070.

18. Friis NF. The pathogenicity of Mycoplasma flocculare. Acta Vet Scand. 1973; 14(2):344–346. PMID: 4728115.
19. Weisburg WG, Tully JG, Rose DL, Petzel JP, Oyalzu H, Yang D, Mandelco L, Sechrest J, Lawrence TG, Van Etten J. A phylogenetic analysis of the mycoplasmas: Basis for their classification. J Bacteriol. 1989; 171(12):6455–6467. PMID: 2592342.
21. Maes D, Verdonck M, Deluyker H, de Kruif A. Enzootic pneumonia in pigs. Vet Quart. 1996; 18(3):104–109.

22. Freeman MJ, Armstrong CH, Freeman-Sands LL, Lopez-Osuna M. Serological cross-reactivity of porcine reference antisera to Mycoplasma hyopneumoniae, M. flocculare, M. hyorhinis and M. hyosynoviae indicated by the enzyme-linked immunosorbent assay, complement fixation and indirect hemagglutination tests. Can J Comp Med. 1984; 48(2):202–207. PMID: 6372971.
23. Armstrong CH, Freeman MJ, Sands-Freeman L. Crossreactions between Mycoplasma hyopneumoniae and Mycoplasma flocculare: practical implications for the serodiagnosis of mycoplasmal pneumonia of swine. Isr J Med Sci. 1987; 23(6):654–656. PMID: 3667232.
24. Cimino GD, Metchette KC, Tessman JW, Hearst JE, Isaaca ST. Post PCR sterilization: a method to control carry-over contamination for the polymerase chain reaction. Nucleic Acids Res. 1991; 19(1):99–107. PMID: 2011516.
25. Porter-Jordan K, Rosenberg EI, Keiser WF. Nested polymerase chain reaction assay for the detection of Cytomegalovirus overcomes false positives caused by contamination with fragmented DNA. J Med Virol. 1990; 30(2):85–91. PMID: 2156009.

26. Levy R, Najioullah F, Thouvenot D, Bosshard S, Aymard M, Lina B. Evaluation and comparison of PCR and hybridization methods for rapid detection of cytomegalovirus in clinical samples. J Virol Methods. 1996; 62(2):103–111. PMID: 9002068.
27. Burns J, Graham AK, Franck C, Fleming KA, Evans MF, McGee JOD. Detection of low copy human papillomavirus DNA and mRNA in routine paraffin sections by non isotopic in situ hybridization. J Clin Pathol. 1987; 40(8):858–864. PMID: 2821078.
28. Syrjanen S, Partanen P, Mantvjarvi R, Syrjanen K. Sensitivity of the in situ hybridization techniques using biotin and 35S labelled human papillomavirus (HPV) DNA probes. J Virol Methods. 1988; 19(3-4):225–238. PMID: 2836460.
29. Cullen AP, Long CD, Lorincz AT. Rapid detection and typing of Herpes Simplex virus DNA in clinical specimens by the hybrid capture II signal amplification probe test. J Clin Microbiol. 1997; 35(9):2275–2278. PMID: 9276401.

30. McClintock JT, Mosher M, Thaker SR, Wacker WK, Jones D, Forman M, Adler SP, Charache P, Taub FE. Culture confirmation of cytomegalovirus and herpes simplex virus by direct enzyme-labeled DNA probes and in situ hybridization. J Virol Methods. 1991; 35(1):81–91. PMID: 1666116.

Figure 1
Gel electrophoresis of amplicons by Mycoplasma hyopneumoniae PCR using consensus primer pairs. Lane 1, Template DNA with 107 pg; 2, Template DNA with 106 pg; 3, Template DNA with 105 pg; 4, Template DNA with 104 pg; 5, Template DNA with 103 pg; 6, Template DNA with 102 pg; 7, Template DNA with 10 pg.
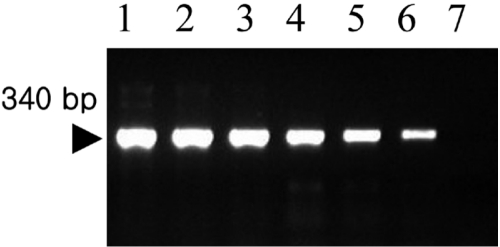
Figure 2
Hybridization was performed with a digoxigenin-labeled mycoplasma consensus DNA probe. Hybrids were detected by an antibody-conjugate (anti-digoxigenin-alkaline phosphatase conjugate) and by a subsequent enzyme-catalyzed color reaction with 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitro blue tetrazolium salt (NBT). 1, Sample DNA with 107 pg; 2, Sample DNA with 106 pg; 3, Sample DNA with 105 pg; 4, Sample DNA with 104 pg; 5, Sample DNA with 103 pg; 6, Sample DNA with 102 pg; 7, Sample DNA with 10 pg.
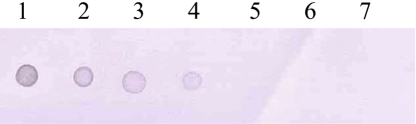
Figure 3
PCR/dot blot hybridization. 1, PCR amplicon of 107 pg; 2, PCR amplicon of 106 pg; 3, PCR amplicon of 105 pg; 4, PCR amplicon of 104 pg; 5, PCR amplicon of 103 pg; 6, PCR amplicon of 102 pg; 7, PCR amplicon of 10 pg.
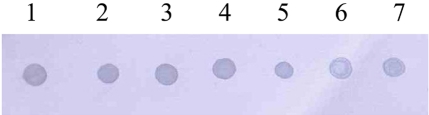
Figure 4
Specificity of PCR/dot blot hybridization. The mycoplasma consensus probe did not react with other pathogens such as Actinobacillus pleuropneumoniae (AP) and Pasteurella multocida (PM). However, PCR/DBH using a Mycoplasma hyopneumoniae (MP) template DNA resulted in a strong positive signal.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download