Abstract
To clone the first anion channel from Xenopus laevis (X. laevis), we isolated a calcium-activated chloride channel (CLCA)-like membrane protein 6 gene (CMP6) in X. laevis. As a first step in gene isolation, an expressed sequence tags database was screened to find the partial cDNA fragment. A putative partial cDNA sequence was obtained by comparison with rat CLCAs identified in our laboratory. First stranded cDNA was synthesized by reverse transcription polymerase-chain reaction (RT-PCR) using a specific primer designed for the target cDNA. Repeating the 5' and 3' rapid amplification of cDNA ends, full-length cDNA was constructed from the cDNA pool. The full-length CMP6 cDNA completed via 5'- and 3'-RACE was 2,940 bp long and had an open reading frame (ORF) of 940 amino acids. The predicted 940 polypeptides have four major transmembrane domains and showed about 50% identity with that of rat brain CLCAs in our previously published data. Semi-quantification analysis revealed that CMP6 was most abundantly expressed in small intestine, colon and liver. However, all tissues except small intestine, colon and liver had undetectable levels. This result became more credible after we did real-time PCR quantification for the target gene. In view of all CLCA studies focused on human or murine channels, this finding suggests a hypothetical protein as an ion channel, an X. laevis CLCA.
The genomic information of primary laboratory animals such as mouse and rat was revealed in the genome project [1,2]. On the basis of those gene constructions in the genome, various gene families were identified for the study of gene function contributing to the scientific needs.
The calcium-activated chloride channel (CaCC) family has been summarized as a group of anion channels that are activated by release of calcium from the endoplasmic reticulum (ER) [3,4]. The CaCC family has at least two types of chloride channels: TMEM16 channels [5-7] and chloride channels calcium-activated (CLCA). Even though this family show a common characteristic of being activated by release of calcium from the ER, there is little genetic and molecular similarity between the two CaCC families [3,4].
Recently, two different CaCCs, xTMEM16A and xANO2, were cloned as Xenopus laevis (X. laevis) CaCCs [8,9]. These two TMEM16 channels, also called anoctamin, have very significant molecular and physiological characteristics. In our research of anoctamin, tissue distribution analysis revealed a ubiquitous expression pattern in various X. laevis tissues including intestines [9].
With a focus on function, calcium-activated chloride channels (CLCAs) have been studied in a number of species including bovine [10], murine [11,12], human [13], and porcine [14]. We reported two CLCAs isolated from one laboratory animal, Ratus norvegicus [15,16]. These rbCLCA1 and rbCLCA2 channels have common characteristics such as primary molecular structure and expression profiles.
As mentioned above, CLCAs from various animal species and tissues have been studied except X. laevis. Moreover, most of the studies were more likely to focus on diseases or functions of humans, and the molecular characterization of the channels tended to be neglected. Here we report a putative CLCA-like membrane protein 6 gene (CMP6) in X. laevis on the basis of its primary molecular structure and tissue distribution profiles.
Chemical reagents were purchased from BioPure (Burlington, Ontario, Canada). pGEM®-T Easy vector was from Promega (Madison, WI, USA). A peltier thermal cycler PTC-200 cycler was purchased from BIO-RAD (Hercules, CA, USA). Female X. laevis (Xenopus I, Ann Arbor, MI, USA) frog was kindly provided by Dr. S.Y. Nah (Konkuk University, Seoul, Korea).
An EST database search was conducted on X. laevis to find out whether there are any partial cDNAs that show high homology with mouse CLCA sequences. First, BLAST server on NCBI was utilized as a search tool to find the X. laevis EST partial cDNA sequence. Highly homologous partial cDNAs were found from EST clones, and among them, a partial cDNA showing 50% nucleotide homology was selected. The amino acid sequence indicated that the sequence was aligned and we compared it to mouse CLCAs (Figure 1A). The amino acid sequence exceeding 45% homology was extracted from a protein database and the intermediate results confirmed that the sequences were significant enough to proceed to the next step. Sequences showing too high or too low homology were not considered candidate sequences and were excluded. Finally, a partial cDNA was selected as a candidate for cloning.
The screening of Xenopus EST database in the NCBI was conducted to query the target sequence. To search for partial cDNAs as candidates for cloning, a partial cDNA (DC066238) was screened. DC066238 cDNA was utilized to generate gene specific primers including C218 for completion of the whole cDNA sequence (Table 1).
Total RNAs were extracted from X. laevis tissues with easy-BLUE (iNtRON Biotechnology, Seongnam, Korea) according to the user's manual with some modifications. To avoid genomic DNA contamination, the extracted total RNA samples were digested with RNase free DNase I (Invitrogen, San Diego, CA, USA) prior to use.
Reverse transcription reactions were performed with 10 µg of total RNA using gene specific primers (10 pmol/µL) and dNTPs. To anneal the RNA and the primers, RNA was heat denatured at 65℃ for 5 min as a conditioning of the RNA-primer mixture. The reaction mixture was as follows: nuclease-free water, 100 mM DTT, RNase inhibitor, and Superscript III RTase, The mixture was incubated at 50℃ for 30 min and at 55℃ for 30 min, and RNA was destroyed by a final concentration of 0.5 M sodium hydroxide. After ethanol precipitation with one-tenth volume of sodium acetate, the reaction product was resuspended in 20 µL of nuclease-free water for 3'-RACE. 5' kination of the cDNA and linker-attachment for the 5'-RACE were performed for effective construction. After all the steps, Purification was done using Sephacryl® S-400 to remove the excessive oligoprimers.
The 5'- and 3'-RACE methods were employed with gene specific primers and an oligo-d(T). These two rounds of RACE reactions were repeated for the amplification of full-length cDNA as shown in Figure 1B. Table 1 represents primer sets used on completion of the two rounds of RACE and construction of a subcloning plasmid. To avoid any match errors of the PCR and RACE reaction, KOD-Plus Taq polymerase (Toyobo, Osaka, Japan) was used through amplification of cDNA. Reaction conditions were as follows: 94℃ for 2 min, 30 cycles of 94℃ for 30 sec, 60℃ for 30 sec, and 68℃ for 1-3 min for the first PCR; for the second PCR, 94℃ for 2 min, 30 cycles of 94℃ for 30 sec, 60℃ for 30 sec, and 68℃ for 1-3 min.
After the two rounds of RACE, Round 1 and Round 2 sub-clones were connected by a recombinant PCR method. Two amplified cDNAs of the 5' and 3' terminuses were used as the overlapping templates together with each piece of DNA to complete a full-length cDNA. C407, C409, and C408 primers were designed from the 5'- and 3'-UTRs. These primer sets for the final step of the recombinant PCR were used for the amplification of full-length cDNA. All these primers are summarized in Table 1. To clone the full-length cDNA, a recombinant PCR product was inserted into a pGEM®-T Easy subcloning vector (Promega). The CMP6 cDNA was confirmed by sequence analysis. For sequence analysis, each cDNA obtained from RACE was tailed with poly-A, ligated with the above vector, and transformed with E. coli DH5α strain. Sequence determination was performed using an ABI PRISM system, and homology alignment and analysis were conducted on an NCBI-BLAST server.
To perform RT-PCR, first cDNA was synthesized from RNA as described in Materials and Methods. Two rounds of PCR were carried out for the 45 repeats to determine the specific products of CMP6. These procedures were carried out in a Peltier thermal cycler PTC-200. For semi-quantification of PCR, nine tissue RNAs (brain, heart, lung, liver, small intestine, colon, kidney, spleen, and oocytes) were employed. In this RT-PCR, i-StarTaq™ polymerase (iNtRON Biotechnology) was used for the stable and fine amplification from nine X. laevis tissue-derived cDNAs with gene specific primers on a Peltier thermal cycler PTC 200. The PCR reaction was performed as follows: pre-denaturation step at 94℃ for 1 min, 20 repeats of denaturation step at 94℃ for 15 sec, annealing at 55℃ for 20 sec, extension at 72℃ for 20℃sec, and for second PCR, 94 for 1 min, 25 repeats of 94℃ for 15 sec, 57℃ for 20 sec, 72℃ for 1 min. Excess primers were removed by Sephacryl® S-400 column chromatography after the first PCR.
To provide a CMP6 quantification result, real-time PCR was performed to confirm whether any difference between RT-PCR and real-time PCR existed. In order to maintain consistency, nine PCR products from reverse-transcription PCR derived from nine same cDNAs were used in the next experiment. A CHROMO4™ (BIO-RAD) real-time PCR system was employed for analysis. DyNAmo™ ColorFlash® SYBR Green qPCR kit (Finnzymes Oy, Espoo, Finland) was utilized with the following steps: pre-denaturation at 95℃ for 7 min, 35 repeats of the denaturation step at 95℃ for 10 sec, at 60℃ for 20 sec, at 72℃ for 20 sec, a final elongation step at 72℃ for 7 min, and establishment of a melting curve from 65 to 90℃. Expression levels were calculated using GeneXpression Macro CHROMO4™ from an Opticon4 program provided by the supplier.
The selected partial cDNA, DC066238, has a 630 bp nucleotide sequence. One ORF codes for a 210 amino acid sequence. This cDNA was screened by comparison of its homology to mouse CLCA1 and CLCA6 which showed over a 48% amino acid similarity (Figure 1A). As a first step in gene cloning, DC066238 was utilized for primer design to clone the CMP6 as described in Materials and Methods.
To complete the full-length CMP6 cDNA, each cDNA, a 1,700 bp linker-C218 (5'-RACE) and a 1,296 bp C401-adapter (3'-RACE) were amplified as described in Materials and Methods (Figure 1B, C, D). It was confirmed that CMP6 had no base changes or missed sequences over the whole region (data not shown). As a result, we have successfully isolated full-length X. laevis CMP6 cDNA including a 2,940 bp sequence with one ORF of 940 amino acids. We did this using several repeats of the 3'- and 5'-RACE method and the specific primers as described in Materials and Methods. The completed CMP6 cDNA contained a short 5'- and a 3'-UTR with a poly (A)n tail and a poly-adenylation signal. We registered this cDNA as a CMP6 gene on the DDBJ database (GenBank Accession No. AB591378).
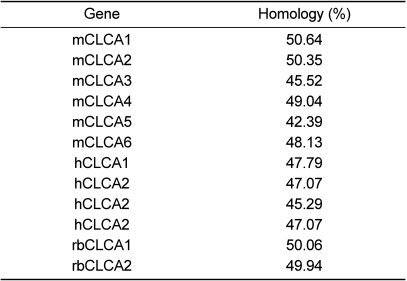
The sequence analysis of the CMP6 gene revealed short 5'- and 3'-UTRs and a single large ORF. The CMP6 protein, including 940 amino acids, was aligned by CLUSTAL W software. The rat CLCAs and CMP6 were compared whether or not it had significant homology or was considered as a CLCA gene family member. The ORF of X. laevis CMP6 showed identity exceeding 50% overall to those of rbCLCA1 (Figure 2A and Table 2). According to this alignment, the overall significant homology that was exhibited led us to regard it as a new Xenopus CLCA-like member (Table 2).
It has been reported that the general CLCAs have four or five transmembrane (TM) domains [17]. Although there are limitations when the actual protein conformation analysis is absent, computational analysis using an appropriate program makes it possible to analyze and predict the conformation of certain protein structures. The hydropathy analysis of CMP6 (Kyte-Doolittle method) indicates that there exist at least four predicted TM domains as shown in Figure 2B. According to hydropathy analysis, we were able to propose that CMP6 is a highly homologous CLCA family member.
To determine the unique characteristics of the CLCA family, the predicted consensus sites were searched for on a GENETYX program. We found the major characteristics of the CLCA family such as the sites of N-linked glycosylation, N-myristoylation, phosphorylation by PKC, casein kinase II and cAMP-dependent protein kinase (Figure 2A). Also, there was a predicted signal sequence and a hydrophobic carboxyl terminus that was regarded as a typical membrane protein including CLCA.
In tissue distribution analysis, the amplified PCR products were detected in some tissues. In particular, we could detect CMP6 gene products, which were expressed predominantly in small intestine, colon and liver (Figure 3). The gene-specific products of CMP6, and β-actin as an internal control, were shown to be 252 and 159 bp (Figure 3A). The intensity of each product was calculated using the E-Capt program for normalization. Relative ratios of CMP6 expression levels in each tissue were calculated compared to the housekeeping gene β-actin (Figure 3B). Each RT-PCR was conducted at least three times to insure the reliability of the numerical values. As shown in Figures 3A and 3B, expression levels were different with small intestine>colon>liver. However, triple repeats of experiments showed that the probability of liver expression is very low (no more than one out of four times; data not shown). In summary, the expression of CMP6 is distinct and specified within the gastrointestinal tract. This suggests that CMP6 expression patterns are limited to secretory functions.
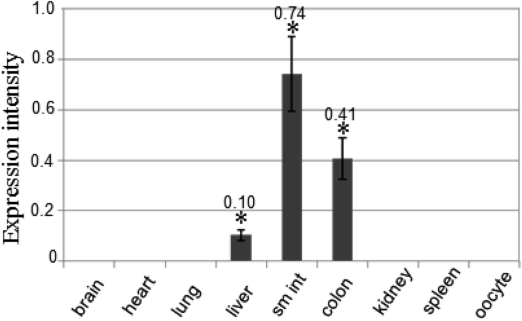
In order to validate data from semi-quantification RT-PCR analysis, quantitative real-time PCR analysis was conducted as described in Materials and Methods. The exponential graph of nine tissues was obtained as the amplification time for expression intensity (data not shown). Figure 4 shows a bar graph that was converted from the exponential graph using numerical values for each product. Expression levels were: small intestine>colon>liver. It also showed that heart was amplified a little (data not shown). The real-time PCR data are completely consistent with the RT-PCR data (Figure 4).
Since there were no reports about the CLCA gene family of X. laevis, we studied X. laevis CLCA gene family members. After two rounds of RACE, linker-C218 constructs were first cloned from X. laevis spleen. However, we could not isolate the exact match of the C218 sequence in spleen tissue. Moreover, there were subtle changes in amino acid sequences. Paradoxically, this kind of difficulty in an experiment suggests the possibility of the existence of a new CLCA gene. Hence, we are now screening for more CLCA members in X. laevis.
In this study, a 2,940 bp cDNA of CMP6 was successfully cloned from X. laevis (Figure 1). As mentioned above, it has been reported that two bovine CLCAs, and five or more murine and human homologs were isolated, but not from X. laevis. Also, two rat brain CLCAs showed lengths around 2,800 bp [15,16]. As regards animal CLCAs, it is meaningful to identify CLCA genes in unstudied animal species. Under homology-based conditions, we can propose that CMP6 may be a functional CLCA. This cDNA was similar to the rat CLCA sequences showing 56% homology for the nucleotide sequence, and a 50% homology for the amino acid sequence [15,16]. There was a low 21% similarity in the restricted partial region of xANO2 [9]. CMP6 shows the most homology (over 50%) to mice CLCA1 and rbCLCA1 (Table 2). Although mouse CLCA1 (mCLCA1) has the highest homology to CMP6, at least if similarities are limited to percentages, rbCLCA1 showed the most homology spanning the overall primary structure (data not shown).
The primary structure of CMP6 revealed four major potential TM domains and, in hydropathy analysis using an analysis window of 17 residues, several minor regions (Figure 2B). As has been proposed for hCLCA1 or hCLCA2, many CLCA family members have four or five TM domains [4]. This analysis is consistent with typical CLCA membrane proteins. On the contrary, two X. laevis anoctamins, xTMEM16A and xANO2, have eight distinct TM domains compared to four TM domains for the CLCA family. These results support the low amino acid sequence homology between CLCAs and anoctamins.
Loewen et al. divided the CLCA gene family into two groups. The first group includes hCLCA1, mCLCA3, and pCLCA1. The second group contains two subgroups - one that contains mCLCA1, mCLCA2, and mCLCA4, and a second that contains hCLCA3, bCLCA1, and bCLCA2 [17]. Evans SR et al. reported two mouse CLCA genes with mCLCA6 expressed primarily in intestine, stomach and in liver a little, whereas mCLCA5 is widely expressed with particularly strong expression in eye and spleen [12]. CMP6 was expressed specifically in intestine (and in liver a little) in contrast to rat CLCAs which were predominantly expressed in brain [15,16].
Moreover, there were similar molecular characteristics regarding the far carboxyl terminus (C-terminus) of the primary structures between CMP6, and mCLCA6 and mCLCA4. This was in contrast to other CLCAs that had no similarity with the C-terminus of CMP6. The C-terminus of CMP6 was most similar to that of mCLCA6 whereas mCLCA4 was less similar (data not shown). Even though we have no evidence for a relation between function and similarities at the C-terminus, we can assume that CMP6 is key in the gastrointestinal tract, especially mucus production. This type of CLCA-like protein might be involved in the regulation of chloride conductance in epithelium with secretory functions. For a more defined study in the future, further work will be needed using heterologous expression analyses.
Acknowledgments
This research was supported by a grant to S. M. Jeong from the Brain Korea 21 project, and a Korean Research Foundation Grant funded by the Ministry of Education & Human Resources Development (KRF-313-2007-2-E00519). This research was supported by the Technology Development Program for Agriculture and Forestry (2010-A008-0034) from the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea. We appreciate a grant supported by the Takara Korea company.
References
1. Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schönbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyooka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y. RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001; 409:685–690. PMID: 11217851.
2. Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera , Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D'Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F. Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004; 428:493–521. PMID: 15057822.
3. Hartzell CH, Putzier I, Arreola J. Calcium-activated chloride channels. Annu Rev Physiol. 2005; 67:719–758. PMID: 15709976.

4. Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005; 85:1061–1092. PMID: 15987802.

5. Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJ. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008; 322:590–594. PMID: 18772398.

6. Huang F, Rock JR, Harfe BD, Cheng T, Huang X, Jan YN, Jan LY. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc Natl Acad Sci USA. 2009; 106:21413–21418. PMID: 19965375.

7. Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shin WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008; 455:1210–1215. PMID: 18724360.

8. Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008; 134:1019–1029. PMID: 18805094.

9. Ryu RH, Oh SJ, Lee RM, Jeong SW, Jan LY, Lee CH, Lee CJ, Jeong SM. Cloning and heterologous expression of new xANO2 from Xenopus laevis. Biochem Biophys Res Commun. 2011; 408(4):559–565. PMID: 21521635.
10. Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. 1995; 270:31016–31026. PMID: 8537359.

11. Evans SR, Thoreson WB, Beck CL. Molecular and functional analyses of two new calcium-activated chloride channel family members from mouse eye and intestine. J Biol Chem. 2004; 279:41792–41800. PMID: 15284223.

12. Lee D, Ha S, Kho Y, Kim J, Cho K, Baik M, Choi Y. Induction of mouse Ca(2+)-sensitive chloride channel 2 gene during involution of mammary gland. Biochem Biophys Res Commun. 1999; 264:933–937. PMID: 10544033.

13. Agnel M, Vermat T, Culouscou JM. Identification of three novel members of the calcium dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett. 1999; 455:295–301. PMID: 10437792.
14. Gaspar KJ, Racette KJ, Gordon JR, Loewen ME, Forsyth GW. Cloning a chloride conductance mediator from the apical membrane of porcine ileal enterocytes. Physiol Genomics. 2000; 3:101–111. PMID: 11015605.

15. Jeong SM, Park HK, Yoon IS, Lee JH, Kim JH, Jang CG, Lee CJ, Nah SY. Cloning and expression of Ca2+-activated chloride channel from rat brain. Biochem Biophys Res Commun. 2005; 334:569–576. PMID: 16023076.
16. Yoon IS, Jeong SM, Lee SN, Lee JH, Kim JH, Pyo MK, Lee BH, Choi SH, Rhim H, Choe H, Nah SY. Cloning and heterologous expression of a Ca2+-activated chloride channel isoform from rat brain. Biol Pharm Bull. 2006; 29:2168–2173. PMID: 17077509.

17. Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999; 59:5488–5491. PMID: 10554024.
Figure 1
CMP6 cDNA isolation. (A) Sequence alignment of DC066238. Amino acid (aa) alignment was deduced by querying the DC066238 amino acid sequence using GENETYX. The amino acid homology is indicated as follows: identical (asterisk), very similar (double dot) and similar (dot). (B) The strategy is shown for generating full-length cDNA. First strand cDNA (dotted) and PCR products (thick) are shown with black lines. CMP6: 5'-RACE and 3'-RACE products represent Round 1 and Round 2, respectively. 3'-RACE products are shown in panel (C) lane 1, and 5'-RACE PCR products in (D) lane 1. PCR products were electrophoresed in a 1% agarose gel. Black arrowheads on panels (C) and (D) indicate the 1,700 and 1,296 bp of target products of CMP6. Molecular weights are shown for 1 kb Plus size markers (G&P, Anyang, Korea).
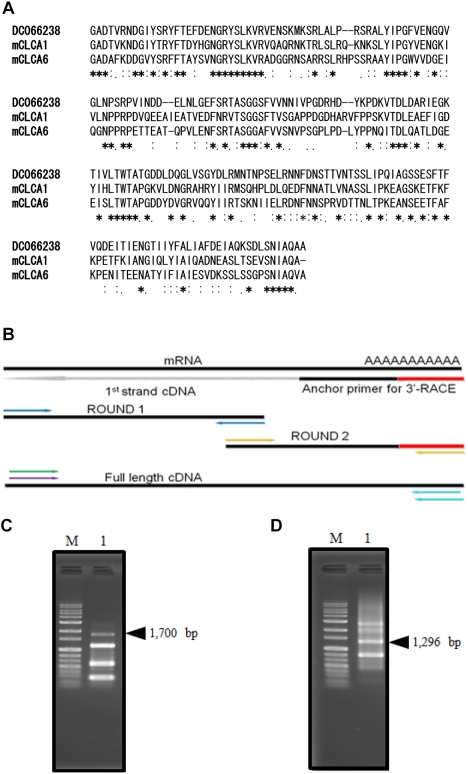
Figure 2
Comparison of X. laevis CMP6 primary structure. (A) CMP6 amino acids were aligned by querying with rat CLCAs on CLUSTAL W. The homology of amino acids of CMP6 is indicated as identical (asterisk) very similar (double dot) and similar (dot). Consensus sites are marked as follows: N-linked glycosylation (●), N-myristoylation sites (▾), phosphorylation by PKC (#), by casein kinase II (◇) and by cAMP-dependent protein kinase (+). The predicted signal sequence (SS) and the hydrophobic carboxyl terminus (H) are overlined. An arrow indicates the predicted cleavage site. (B) Hydropathy analysis. The hydropathy plot was analyzed by the Kyte-Doolittle method. Hydrophobic domains are given as positive values. The proposed hydrophobic domains are four segments.
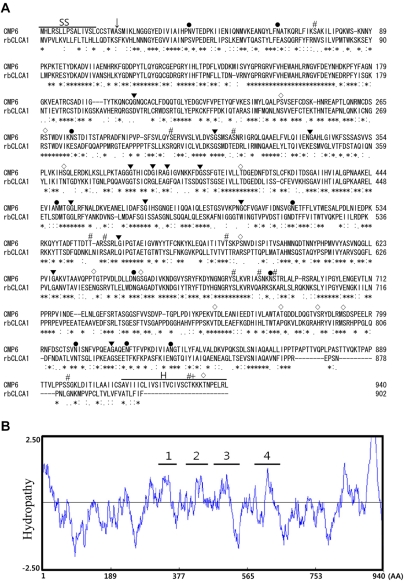
Figure 3
Tissue distribution of CMP6 expression in X. laevis. (A) RT-PCR products in various tissues. The amplified PCR products were electrophoresed in a 2% agarose gel. The specific RT-PCR products are a 252 bp CMP6 (C415 and C416) and a 159 bp β-actin gene as an internal control (XHRMbA1 and xbA3). Expression ratios of CMP6 are shown as relative calculated values for β-actin. (B) CMP6 showed high expression levels in small intestine intestine (sm int) and colon. Each graph represents at least three independent experiments. The bars shown in the graph are mean±SD. *P<0.05 compared to control group.
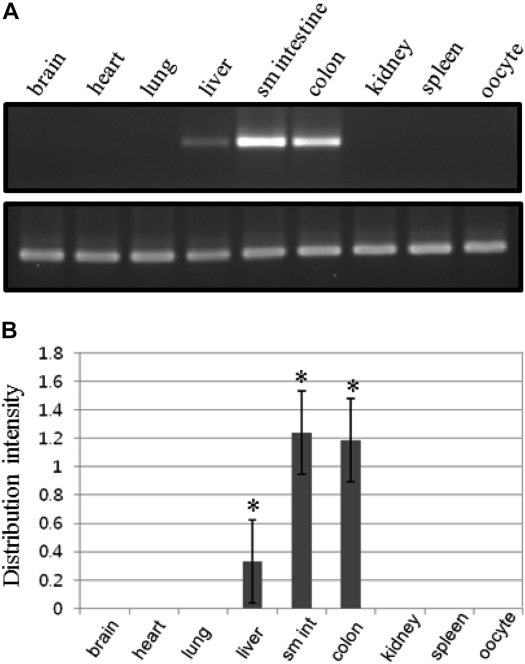
Figure 4
CMP6 quantification by real-time PCR. Relative quantification is shown as a bar graph. The relative CMP6 expression ratios for β-actin are shown. This result confirms the idea of CMP6 being largely expressed in liver, small intestine (sm int), and colon. The graph represents three independent experiments with each bar as mean±SD. *P<0.05 compared to control group.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download