Abstract
Asthma exacerbation (AE) usually denotes worsening of asthma symptoms that requires intense management to prevent further deterioration. AE has been reported to correlate with clinical and demographic factors, such as race, gender, and treatment compliance as well as environmental factors, such as viral infection, smoking, and air pollution. In addition, recent observations suggest that there are likely to be genetic factors specific to AE. Understanding genetic factors specific to AE is essential to develop therapy tailored for exacerbation-prone asthma. Here, we summarize the results of studies involving genetic risk factors for AE. To simplify and enhance understanding, we reviewed the studies according to the following categories: hypothesis-driven approaches, hypothesis-free approaches, gene-environment interactions, and pharmacogenetics.
Asthma is a chronic inflammatory disorder characterized by variable airway obstruction. Occasionally, patients with asthma experience sudden deteriorations of their symptoms which are referred to as an ‘exacerbation.’ Although there are diverse viewpoints, asthma exacerbation usually denotes worsening of asthma symptoms that requires intense management to prevent further deterioration. For instance, in the National Institute of Health-sponsored Asthma Outcomes workshop, asthma exacerbation was defined as worsening of asthma requiring the use of systemic corticosteroids to prevent a serious outcome.1 Exacerbations have been reported to correlate with clinical and demographic factors, such as race, gender, and treatment compliance as well as environmental factors, such as viral infection, smoking and air pollution.2345678 To reduce the risk of asthma exacerbation (AE), a tailored application of standard treatment based on detailed phenotyping was proposed.9
A complex interaction between genetic and nongenetic factors has been thought to be important in asthma pathogenesis. Thus, it is not surprising that heritability estimates have indicated that, on average, genetic variation among individuals accounts for one-half of the risk of asthma.10 Interestingly, recent observations show that genetic risk factors important in asthma progression and severity differ from those that determine susceptibility,11 which suggests that there must be genetic factors specific to AE. For example, genetic predictors of a poor long-term response to inhaled corticosteroids (ICSs) markedly differ according to outcomes (exacerbations vs lung function) in childhood asthmatics.12 Understanding genetic factors specific to AE is essential to develop specific therapy tailored for exacerbation-prone asthma. With this in mind, we aimed to summarize genetic association studies mainly focused on AE in this review. Here, a severe AE represents acute symptoms requiring emergency department visit, hospitalization, and intubation; a less severe AE represents acute symptoms requiring treatment with an oral or parenteral corticosteroid. To simplify and enhance understanding, we reviewed the studies according to the following categories: hypothesis-driven or candidate gene approaches, hypothesis-free or genome-wide approaches, gene-environment interactions, and pharmacogenetics.
Although asthma is currently believed to be a heterogeneous disease, an allergic asthma characterized by Th2-type T cells, IgE, eosinophils, mast cells, and cytokines is a traditional concept in terms of immunopathology.13 The early and pioneering studies driven by a prior hypothesis focused on this Th2 asthma phenotype. A previous report using a murine model of asthma showed that IL-13 is a critical cytokine in the development of asthma.14 For this reason, genes related with the IL-13 signaling pathway have been frequent targets of asthma genetic studies. Hunningghake et al.15 reported that rs1800925, a functional single nucleotide polymorphism (SNP) located at the promoter area of IL-13, is associated with the risk of severe AEs among Costa Rican and non-Hispanic children with asthma. Both IL-13 and IL-4 signals act via the common IL-4 receptor alpha (IL-4Rα).16 In 2 well-characterized cohorts of severe asthmatics, 2 non-synonymous coding SNPs (rs1805011 [E375A] and rs-1801275 [Q551R])were associated with AE requiring an intensive care unit stay or intubation in addition to lower lung function and mast cell-related tissue inflammation.17
The gene CHI3L1 coding for the protein chitinase 3-like-1 (CHI3L1) is reported to be a susceptibility gene for asthma18 and CHI3L1 is thought to be a possible biomarker for asthma and decline in lung function. Based on this finding, a cross-sectional survey of 1,071 children and young adults with asthma (aged 3 to 22 years) was performed in Scotland.19 They showed that rs4950928 which is associated with CHI3L1 levels significantly increases the risk of severe AEs.19 However, in this study, investigators could not find any significant association between this SNP and less severe AEs defined as oral steroid intake attributable to AEs.19 A prominent candidate is the loci on chromosome 17q12-q21 identified as an asthma susceptibility locus from a genome-wide association study (GWAS).20 In this context, rs7216389 in this region is an eQTL SNP of ORMDL3 and was associated with approximately 2-fold increased risk of AEs in a prospective birth cohort.21 The increased risk of exacerbations persisted from 1 to 6 years of age.21 Interestingly, in some studies, significant associations were observed only in specific groups (e.g. rs1800925 in asthmatics on inhaled corticosteroid15 and rs1805011 and rs1801275 in African American16), which suggests that AE risk genes should be searched in consideration of ethnicity, treatment, environment, and so on. The results of the studies using a hypothesis-driven approach are summarized in Table 1.
A hypothesis-free approach means a genome screening without any preexisting predilection for specific areas, genes, or variants22 and has provided a comprehensive description of the genetic causes of complex disease. A recent GWAS performed in childhood asthmatics in Denmark has identified 5 loci with genome-wide significances associated with severe AEs occurring between 2 and 6 years.23 Among those 5 loci, CDHR3 (encoding cadherin-related family member 3) was not previously reported as asthma susceptibility loci. Discovery of a new AE risk gene is partly attributable to the fact that this study focused on a specific asthma phenotype characterized by recurrent and severe AE, which implies the strength of applying specific phenotyping in asthma genetic studies. Another GWAS focusing on less severe AEs identified several variants in CTNNA3 and SEMA3D as risk loci in 2 independent pediatric asthma clinical trials.24 Among them, rs10997296 in CTNNA3 locus with a genome-wide significance was associated with CTNNA3 mRNA expression in CD4+ cells from asthmatic patients and rs993312 in SEMA3D locus replicated in a clinical biobank database of childhood and young adult asthmatics, though its significance did not reach a genome-wide significance in the primary analysis.24
As mentioned before, complex and multiple traits are associated with AE. A lack of statistical power which comes from small effects of many causal factors, possible interactions among the causal factors, and small sample sizes is an important challenge for prediction of AE, a complex trait. Although overfit pitfalls exist, development of a predicting model using data mining methods is a way to handle this type of problems.25 Random Forests (RF) are a classification algorithm that is composed of a set of random decision trees,2627 and we can integrate hundreds of SNPs and clinical traits to predict complex clinical phenotypes by RF. Xu et al.28 performed a GWAS of severe AE in a pediatric asthma clinical trial and identified a list of top GWAS SNPs based on RF importance score. The authors predicted severe AEs using the top 10 to 320 SNPs together with age, sex, prebronchodilator FEV1 percentage predicted, and treatment group. With only 417 childhood asthmatics, using 160 SNPs, they could generate a good predictive model for severe AEs, with a >0.66 area under the curve and about 0.66 sensitivity and 0.6 specificity. A hypothesis-free approach can help us search unknown genetic markers associated with AEs. In addition, by using machine learning methods, we can increase our understanding of the basis for epistatic (gene-gene) interactions underlying AEs. The results of the studies using a hypothesis-free approach are summarized in Table 2.
In the study by Xu et al.,28 using the clinical traits alone in the predictive model yielded an area under the curve score of 0.54, which suggests that AE is affected by non-genetic as well as genetic factors. Among non-genetic factors, indoor and outdoor environment exposures pose an important influence on AE.29 A gene and environment interaction is defined as a different effect of a genotype on disease risk in persons with different environmental exposures.30 A search for genetic factors showing significant interactions with environmental exposures is clinically significant as environmental exposures may be modifiable. For indoor exposures, rs512625 in ADAM33 located in chromosome 20p13 showed significant interactive effects with environmental tobacco smoke on severe AE.31 High mold exposure (≥25,000 units per gram of house dust) significantly modified the relation between 3 SNPs in CHIT1 (rs2486953, rs4950936, and rs1417149) and severe AE.32 Endotoxin inhalation can aggravate symptoms by causing bronchial obstruction among patients with established asthma.33 Two genetic variants (rs2915863 in CD14 and rs17226566 in LY96) in the endotoxin signaling pathway showed different relations between environment endotoxin exposure and severe AE, respectively, according to their genotypes.34 An exposure to allergen is also one of the important factors contributing to AE. Sharma et al.35 reported that dust mite exposure is significantly modified the effect of rs2241712 in TGFB1 on severe AE. All these studies were performed in childhood asthmatics, and thus interactive effects between genetic factors and indoor environment exposures in adults asthmatics are need to be further investigated. Most studies on the interaction between genetic predispositions and air pollutants (outdoor environmental exposures) focused on pulmonary functions and asthma susceptibility, and none did focus on AE. For this concise review, we did not cover studies searching associations between epigenetic factors and occupational exposures in this manuscript.
Viral respiratory tract infections are known to be an important risk factor for AE. Bosco et al.36 followed exacerbation-prone asthmatic children prospectively and evaluated differential gene expressions in nasal lavage samples obtained during an acute, moderate, picorna virus-induced exacerbation, and 7 to 14 days later. They found that more than 1,000 genes were upregulated during AEs in comparison with 7 to 14 days later and revealed that interferon regulatory factor 7 was one of the hyperconnected hub nodes using coexpression network analysis. The interferon regulatory factor 7 gene is an important regulator of antiviral immunity.37 Interestingly, a recent comprehensive review showed evidence of individual susceptibility to viral infection.5 Taken together, genetic predispositions may play a role in virus-induced AE. In this respect, CDHR3 which was identified as a gene associated with AE in a recent GWAS23 is of particular interest. CDHR3 is a transmembrane protein and highly expressed in the human lung and airway epithelium.38 Recently, it was reported that CDHR3 acts as an rhinovirus C receptor.39 In addition, introduction of the risk variant of rs6967330 in CDHR3 resulted in 10-fold enhanced rhinovirus C binding.39 These findings suggest that CDHR3 variants may confer risk to severe AE via an interaction with rhinovirus C infection. These findings may also explain why CDHR3 was not previously reported as asthma susceptibility loci and has now been identified only in a GWAS focusing on a specific phenotype, severe AE. A hypothesis-free approach is also applicable to the gene and environment interaction analysis. Du et al.40 conducted a GWAS of gene-vitamin D interaction on AEs in participants in the Childhood Asthma Management Program. They identified 3 common variants in the class I MHC-restricted T cell–associated molecule gene (CRTAM) that were associated with an increased rate of AEs in subjects with a low circulating vitamin D level. Results were replicated in a second independent Costa Rican cohort. As genome-wide gene-environment interaction analyses are often underpowered after adjustment for multiple comparisons, Sordillo et al.41 utilized differential gene expression in response to the environmental exposure to capture the most relevant genes. The top differentially expressed genes, IL9, IL5, and PRG2 (proteoglycan 2), were identified through comparison genome-wise gene expression on PBMCs from childhood asthmatics between pre-and post stimulation of dust mite allergen (Der f 1) for 72 hours. They then evaluated whether the 3 genes showed significant interactive effects with home Der p 1 allergen levels on severe AEs in 2 independent cohorts of childhood asthmatics. They found that subjects with the dominant genotype for these IL9 polymorphisms (rs11741137, rs2069885, and rs1859430) were more likely to report a severe AE if exposed to increased dust mite levels. The results of the studies showing gene-environment interactions are summarized in Table 3.
The current primary therapies for asthma are focused on increasing bronchial smooth muscle relaxation (bronchodilation) and decreasing airway inflammation. The response to drug treatment in asthma is a complex trait and markedly variable even in patients with apparently similar clinical features.42 In asthma, a poor response to drug treatment is related to AEs.43 Pharmacogenetics is usually defined as a study of variations of DNA characteristics as related to drug response.44 AE is one of the important target phenotypes in pharmacogenetic studies of asthma. Inhaled β2-agonists are widely used in asthma treatment for the purpose of bronchodilation and act via their binding to the β2 adrenergic receptor (β2AR). The effect of genetic variants in the ADRB2 coding β2AR on receptor function has been intensively examined.4546 For this reason, pharmacogenetics studies on heterogeneity in response to inhaled β2-agonists have mostly focused on ADRB2. A recent meta-analysis stratified by treatment showed that the use of a long acting β2-agonist (LABA), but not an leukotriene receptor antagonist, as an add-on controller is associated with increased risk of AE in children carrying 1 or 2 A alleles at rs1042713 (Arg16 amino acid) in ADRB2.47 In this study, data from 4,226 children of white Northern European and Latino origin was analyzed, and both severe and non-severe AEs were included. However, a pharmacogenetic analysis of 2 prospective clinical trials undertaken in adult asthmatics showed no effect of ADRB2 variation, including rs1042713, on severe AE when they were treated with LABA plus (ICS).48 This difference suggests that there might be a specific genetic mechanism regulating AE in response to a LABA in children that does not carry over to adults. Although ICS is the main stay of asthma treatment, individual responses vary widely and some asthmatics experience exacerbations while on ICS.49 The SNP in FCER2 (T2206C) rs28364072 was reported to be associated with severe AEs in both white and African American children with asthma enrolled in the CAMP trial.50 This strong association demonstrated an almost 4-fold increased risk of exacerbations for subjects homozygous for the mutant allele and was replicated in asthmatic children of European origin.51 Two SNPs in ST13 (rs138335 and rs138337), coding for a co-chaperone of the glucocorticoid receptor, were associated with AEs in the previous year in asthmatic children and young adults despite their ICS use.52 In addition, P2RX7 were identified as a gene associated with AEs in patients taking ICS via a hypothesis-driven approach.53 A GWAS of 237,726 common, independent markers which was conducted in 806 Caucasian asthmatic patients reveled that 6 novel SNPs associated with differential risk of severe AEs.54 Among them, 2 SNPs (rs2395672 in CMTR1 and rs279728) were associated with increased risk of AEs, while 4 SNPs (rs4271056, rs6467778, rs2691529, and rs9303988) were associated with decreased risk of AEs. Two SNPs (rs730012 in LTC4S and rs2660845 in LTA4H) were reported to be associated with AEs in adult asthmatics treated by montelukast for 6 months.55 Currently, biological drugs targeting specific cytokines are under active development with a hope that these drugs would be helpful for severe asthmatics with frequent AEs.56 Pitrakinra, which acts as an antagonist at the heterodimeric receptor complex (IL-4Rα/IL-13Rα1) shared by both IL-4 and IL-13, is a good example.57 Interestingly, it is reported that pitrakinra shows a dose-response relationship for AEs according to the genotype of SNPs that tagged blocks of correlated IL4RA polymorphisms in non-Hispanic white adults with asthma.58 All these findings clearly suggests that pharmacogenetics may be useful to identify an asthma subgroup that is more responsive to therapy and thus reduce the possibility of AEs. The results of pharmacogenetic studies are summarized in Table 4.
The goal of asthma treatment is to prevent recurrent exacerbations. However, we do not have reliable biomarkers predicting in what patient and when AEs will occur. A search for such biomarkers is very important in that new therapies that exhibit relatively selective benefits in reducing AEs become available. Accumulated evidence now suggests the potential role of genetic variants in determining the risk for AE. Figure summarizes plausible biological mechanisms underlying genes associated with asthma exacerbation. However, genetic factors associated with susceptibility to asthma and those associated with frequency of AEs are often inseparably related. Thus, it is possible that some genes listed here could be rather specific to particular endotypes into asthma which are prone to frequent AE. Future investigations of the effects of genetic variants upon AE should consider the interactions of genes with race/ethnicity, medication use, and atopic status as well as previously reported environmental factors. In addition, integrated approaches based on recent advances in omics technology need to be utilized in future study on the association between genes and AEs.
Figures and Tables
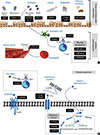 | FigureSummary of plausible biological mechanisms underlying genes associated with asthma exacerbation. (A) Genes related with gene-environment interactions and asthma pathogenesis. (B) Genes related with pharmacogenetic effects. β2AR, β2-adrenergic receptors; CysLTR1, cysteinyl leukotriene receptor 1; GC, glucocorticoid; GR, glucocorticoid receptor; LTC4, leukotriene C4. |
Table 1
Studies using a hypothesis-driven approach

Table 2
Studies with a hypothesis-free approach

Table 3
Studies showing gene-environment interactions

Table 4
Pharmacogenetic studies

References
1. Fuhlbrigge A, Peden D, Apter AJ, Boushey HA, Camargo CA Jr, Gern J, et al. Asthma outcomes: exacerbations. J Allergy Clin Immunol. 2012; 129:S34–S48.
2. Rumpel JA, Ahmedani BK, Peterson EL, Wells KE, Yang M, Levin AM, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol. 2012; 130:1302–1306.
3. Moorman JE, Akinbami LJ, Bailey CM, Zahran HS, King ME, Johnson CA, et al. National surveillance of asthma: United States, 2001-2010. Vital Health Stat 3. 2012; (35):1–58.
4. Williams LK, Peterson EL, Wells K, Ahmedani BK, Kumar R, Burchard EG, et al. Quantifying the proportion of severe asthma exacerbations attributable to inhaled corticosteroid nonadherence. J Allergy Clin Immunol. 2011; 128:1185–1191.e2.
5. Busse WW, Lemanske RF Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet. 2010; 376:826–834.
6. Dulek DE, Peebles RS Jr. Viruses and asthma. Biochim Biophys Acta. 2011; 1810:1080–1090.
7. Chilmonczyk BA, Salmun LM, Megathlin KN, Neveux LM, Palomaki GE, Knight GJ, et al. Association between exposure to environmental tobacco smoke and exacerbations of asthma in children. N Engl J Med. 1993; 328:1665–1669.
8. Samoli E, Nastos PT, Paliatsos AG, Katsouyanni K, Priftis KN. Acute effects of air pollution on pediatric asthma exacerbation: evidence of association and effect modification. Environ Res. 2011; 111:418–424.
9. Custovic A, Johnston SL, Pavord I, Gaga M, Fabbri L, Bel EH, et al. EAACI position statement on asthma exacerbations and severe asthma. Allergy. 2013; 68:1520–1531.
10. Ober C. Asthma genetics in the post-GWAS era. Ann Am Thorac Soc. 2016; 13:Suppl 1. S85–S90.
11. Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol. 2012; 130:861–868.e7.
12. Rogers AJ, Tantisira KG, Fuhlbrigge AL, Litonjua AA, Lasky-Su JA, Szefler SJ, et al. Predictors of poor response during asthma therapy differ with definition of outcome. Pharmacogenomics. 2009; 10:1231–1242.
13. Holgate ST. Pathogenesis of asthma. Clin Exp Allergy. 2008; 38:872–897.
14. Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998; 282:2258–2261.
15. Hunninghake GM, Soto-Quirós ME, Avila L, Su J, Murphy A, Demeo DL, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007; 120:84–90.
16. Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999; 17:701–738.
17. Wenzel SE, Balzar S, Ampleford E, Hawkins GA, Busse WW, Calhoun WJ, et al. IL4R alpha mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med. 2007; 175:570–576.
18. Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008; 358:1682–1691.
19. Cunningham J, Basu K, Tavendale R, Palmer CN, Smith H, Mukhopadhyay S. The CHI3L1 rs4950928 polymorphism is associated with asthma-related hospital admissions in children and young adults. Ann Allergy Asthma Immunol. 2011; 106:381–386.
20. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007; 448:470–473.
21. Bisgaard H, Bønnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Møller E, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009; 179:179–185.
22. Kitsios GD, Zintzaras E. Genome-wide association studies: hypothesis-“ free” or “engaged”? Transl Res. 2009; 154:161–164.
23. Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014; 46:51–55.
24. McGeachie MJ, Wu AC, Tse SM, Clemmer GL, Sordillo J, Himes BE, et al. CTNNA3 and SEMA3D: Promising loci for asthma exacerbation identified through multiple genome-wide association studies. J Allergy Clin Immunol. 2015; 136:1503–1510.
25. Czika WA, Weir BS, Edwards SR, Thompson RW, Nielsen DM, Brocklebank JC, et al. Applying data mining techniques to the mapping of complex disease genes. Genet Epidemiol. 2001; 21:Suppl 1. S435–S440.
26. Breiman L. Random forests. Mach Learn. 2001; 45:5–32.
27. Banfield RE, Hall LO, Bowyer KW, Kegelmeyer WP. A comparison of decision tree ensemble creation techniques. IEEE Trans Pattern Anal Mach Intell. 2007; 29:173–180.
28. Xu M, Tantisira KG, Wu A, Litonjua AA, Chu JH, Himes BE, et al. Genome wide association study to predict severe asthma exacerbations in children using random forests classifiers. BMC Med Genet. 2011; 12:90.
29. Etzel RA. How environmental exposures influence the development and exacerbation of asthma. Pediatrics. 2003; 112:233–239.
30. Ottman R. Gene-environment interaction: definitions and study designs. Prev Med. 1996; 25:764–770.
31. Bukvic BK, Blekic M, Simpson A, Marinho S, Curtin JA, Hankinson J, et al. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr Allergy Immunol. 2013; 24:10–18.
32. Wu AC, Lasky-Su J, Rogers CA, Klanderman BJ, Litonjua AA. Fungal exposure modulates the effect of polymorphisms of chitinases on emergency department visits and hospitalizations. Am J Respir Crit Care Med. 2010; 182:884–889.
33. Michel O, Duchateau J, Sergysels R. Effect of inhaled endotoxin on bronchial reactivity in asthmatic and normal subjects. J Appl Physiol (1985). 1989; 66:1059–1064.
34. Kljaic-Bukvic B, Blekic M, Aberle N, Curtin JA, Hankinson J, Semic-Jusufagic A, et al. Genetic variants in endotoxin signalling pathway, domestic endotoxin exposure and asthma exacerbations. Pediatr Allergy Immunol. 2014; 25:552–557.
35. Sharma S, Raby BA, Hunninghake GM, Soto-Quirós M, Avila L, Murphy AJ, et al. Variants in TGFB1, dust mite exposure, and disease severity in children with asthma. Am J Respir Crit Care Med. 2009; 179:356–362.
36. Bosco A, Ehteshami S, Panyala S, Martinez FD. Interferon regulatory factor 7 is a major hub connecting interferon-mediated responses in virus-induced asthma exacerbations in vivo. J Allergy Clin Immunol. 2012; 129:88–94.
37. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005; 434:772–777.
38. Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005; 21:650–659.
39. Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015; 112:5485–5490.
40. Du R, Litonjua AA, Tantisira KG, Lasky-Su J, Sunyaev SR, Klanderman BJ, et al. Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect asthma exacerbations. J Allergy Clin Immunol. 2012; 129:368–373.
41. Sordillo JE, Kelly R, Bunyavanich S, McGeachie M, Qiu W, Croteau-Chonka DC, et al. Genome-wide expression profiles identify potential targets for gene-environment interactions in asthma severity. J Allergy Clin Immunol. 2015; 136:885–892.e2.
42. Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002; 109:410–418.
43. Greenberg S. Asthma exacerbations: predisposing factors and prediction rules. Curr Opin Allergy Clin Immunol. 2013; 13:225–236.
44. Park HW, Tantisira KG, Weiss ST. Pharmacogenomics in asthma therapy: where are we and where do we go? Annu Rev Pharmacol Toxicol. 2015; 55:129–147.
45. Green SA, Turki J, Bejarano P, Hall IP, Liggett SB. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 1995; 13:25–33.
46. Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000; 97:10483–10488.
47. Turner S, Francis B, Vijverberg S, Pino-Yanes M, Maitland-van der Zee AH, Basu K, et al. Childhood asthma exacerbations and the Arg16 β2-receptor polymorphism: A meta-analysis stratified by treatment. J Allergy Clin Immunol. 2016; 138:107–113.e5.
48. Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007; 370:2118–2125.
49. Bateman ED, Harrison TW, Quirce S, Reddel HK, Buhl R, Humbert M, et al. Overall asthma control achieved with budesonide/formoterol maintenance and reliever therapy for patients on different treatment steps. Respir Res. 2011; 12:38.
50. Tantisira KG, Silverman ES, Mariani TJ, Xu J, Richter BG, Klanderman BJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007; 120:1285–1291.
51. Koster ES, Maitland-van der Zee AH, Tavendale R, Mukhopadhyay S, Vijverberg SJ, Raaijmakers JA, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy. 2011; 66:1546–1552.
52. Vijverberg SJ, Koster ES, Tavendale R, Leusink M, Koenderman L, Raaijmakers JA, et al. ST13 polymorphisms and their effect on exacerbations in steroid-treated asthmatic children and young adults. Clin Exp Allergy. 2015; 45:1051–1059.
53. Denlinger LC, Manthei DM, Seibold MA, Ahn K, Bleecker E, Boushey HA, et al. P2X7-regulated protection from exacerbations and loss of control is independent of asthma maintenance therapy. Am J Respir Crit Care Med. 2013; 187:28–33.
54. Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE, et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis. 2015; 3:350–359.
55. Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006; 173:379–385.
56. Pelaia G, Vatrella A, Maselli R. The potential of biologics for the treatment of asthma. Nat Rev Drug Discov. 2012; 11:958–972.
57. Burmeister Getz E, Fisher DM, Fuller R. Human pharmacokinetics/ pharmacodynamics of an interleukin-4 and interleukin-13 dual antagonist in asthma. J Clin Pharmacol. 2009; 49:1025–1036.
58. Slager RE, Otulana BA, Hawkins GA, Yen YP, Peters SP, Wenzel SE, et al. IL-4 receptor polymorphisms predict reduction in asthma exacerbations during response to an anti-IL-4 receptor α antagonist. J Allergy Clin Immunol. 2012; 130:516–522.e4.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download