Abstract
Purpose
Interleukin-1 (IL-1) plays a key role in inflammation and immunity and its decoy receptor, IL-1R2, has been implicated in transcriptomic and genetic studies of asthma.
Methods
Two large asthma family collections, the French-Canadian Saguenay—Lac-St-Jean (SLSJ) study and the French Epidemiological Study on the Genetics and Environment of Asthma (EGEA), were used to investigate the association of SNPs in 10 genes that modulate IL-1R2 activities with asthma, allergic asthma, and atopy. Gene-gene interactions were also tested.
Results
One SNP in BACE2 was associated with allergic asthma in the SLSJ study and replicated in the EGEA study before statistical correction for multiple testing. Additionally, two SNPs in the MMP2 gene were replicated in both studies prior to statistical correction and reached significance in the combined analysis. Moreover, three gene-gene interactions also survived statistical correction in the combined analyses (BACE1-IL1RAP in asthma and allergic asthma and IL1R1-IL1RAP in atopy).
Over 300 genes related to the asthma phenotype have been identified in genetic association studies.1 However, some genes and loci have been associated, replicated numerous times, and are recognized as the "gold standard". Several association studies have reported the 2q12 locus association with asthma.2 Among the genes of this locus, interleukin 1 (IL1) receptor type 1 (IL1R1) has been associated with innate immunity and the IL1 receptor type 2 (IL1R2) with atopy.34 IL1R2 was also found to be overexpressed in bronchial biopsies of individuals with allergic asthma.5
IL-1 is a potent cytokine of innate immunity primarily involved in the initiation of inflammation, as well as Th17 and nonconventional Th1 (CD161+) responses. It has been shown that drugs neutralizing IL-1 lead to a reduction of airway hyperreactivity, a decrease of inflammatory infiltration and less Th2 cytokines.678 IL-1 has two receptors: 1) IL-1R1 is expressed on virtually all cell types; and 2) IL-1R2 is principally expressed by neutrophils, B-cells, monocytes, and macrophages, as well as in the plasma in its soluble form (sIL-1R2).9 IL-1R2 is involved in the negative regulation of IL-1 as it does not possess the Toll-like domain essential for signal transduction10 and sIL-1R2 can bind directly to IL-1 and sIL-1RAP.9 As the IL1R1 and IL1R2 genes have been implicated in asthma, we hypothesized that genes that modulate IL1R2 activity could have a role in asthma and asthma-related phenotypes. To investigate this hypothesis, we selected single nucleotide polymorphisms (SNPs) of 10 genes involved in IL1R2 activities and investigated their associations with asthma, allergic asthma, and atopy using 2 familial collections.
The French-Canadian Saguenay—Lac-St-Jean (SLSJ) asthma familial collection and the French Epidemiological Study on the Genetics and Environment of Asthma (EGEA) are both large collections that bring power to this study. The SLSJ familial collection is comprised of 1,284 individuals (253 unrelated asthmatic probands) native of SLSJ, a region of Québec (Canada) and was used in the discovery study.11 This population of European origin is known for its founder effect and has a wide range of highly conserved chromosomal regions, facilitating the identification of disease-associated genes. A total of 789 individuals (235 probands) with European ancestry12 from the EGEA family collection were included in this study and used for replication. All probands and their two parents were of European ancestry, born in France.12 A description of the studies and a table describing the phenotypic features are available in the online supplement (Table E1). For SLSJ and EGEA independent studies, ethical approval was obtained from the appropriate institutional ethic committees and all individuals gave written informed consent.
The power to detect single SNP associations and SNP × SNP interactions in the discovery collection was determined using the Quanto program (http://biostats.usc.edu/Quanto.html).1314 The power was estimated under an additive genetic model with minor allele frequencies (maf) between 0.05 and 0.50 and main effect size (odds ratio) or interaction effect size (odds ratio in interaction analyses) ranging from 1.1 to 2.5. The study had sufficient power (>80%) to detect moderate to large effects (≥1.7) for SNPs with maf ≥0.05 and sufficient power to detect small to moderate effects (≥1.4) for SNPs with maf ≥0.20 in single SNP analyses. For the interaction analyses, the study had sufficient power to detect moderately large effects between common alleles (odds ratio ≥1.7 for maf ≥0.15 and an odds ratio ≥1.4 for maf ≥0.30). Full details of the power calculations are in Tables E2 and E3.
To identify genes involved in IL1R2 function, a review of the literature was performed using PubMed by entering different combinations of the following keywords: IL1R2, interleukin 1 decoy receptor, expression, induce, regulate, decrease, and increase. A total of 10 genes were selected according to their direct effect on the IL1R2 expression or protein level or for the direct impact on IL-1R2 binding activity. The genes are presented in Figure and Supplementary Table E4.
Genotypes were extracted from the genome-wide data obtained in the context of the European GABRIEL asthma consortium using the 610K Quad Array from Illumina (Illumina, San Diego, CA, USA). A single SNP analysis was performed using the Family-Based Association Test (FBAT; http://www.hsph.harvard.edu/fbat/default.html) under an additive genetic model. Analyses of gene-gene interactions including 17,040 SNP pairs were tested using UNPHASED software (v3.1.5) (https://sites.google.com/site/fdudbridge/software/unphased-3-1) with the «genotype tests» option. The most frequent haplotype for each pair of SNPs was used as the reference haplotype for the interactions. Moreover, only haplotypes with a frequency ≥1% were examined. Analyses were first performed in the discovery SLSJ family dataset, and the most significant signals were used for replication in EGEA families (P≤0.05 for single SNPs and P≤0.0005 for interaction analyses (corrected P value considering the number of independent SNPs). Using the SNPSpD and the matSpD tools, the critical P value thresholds for single SNP analysis was set to P≤0.0003. Using a Bonferroni correction, the statistical threshold was set to P≤2.9×10-6 for the SNP×SNP interactions (correcting for the number of SNP × SNP interactions and independent phenotypes).15 For the replication analysis, the statistical thresholds were obtained for each phenotype according to the number of SNPs tested and were set to P≤0.0048, P≤0.0030, and P≤0.0056 for asthma, allergic asthma and atopy phenotypes, respectively in single SNP analyses. The thresholds were set to P≤0.05 for the SNP×SNP interaction analyses for all phenotypes as only one different association was tested for each. These statistical thresholds were also applied to the combined meta-analysis results of both the discovery and replication studies. SLSJ and EGEA results were combined using the Stouffer's method implemented in METAL (single SNP analyses; http://www.sph.umich.edu/csg/abecasis/metal/) and MetaP software (interaction analyses; http://compute1.lsrc.duke.edu/softwares/MetaP/metap.php).
A total of 184 SNPs were located in a±10 kb region of the 10 selected genes and met the following quality control criteria (call rate ≥95%; maf ≥5%; Hardy-Weinberg equilibrium P value ≥5%; and Mendelian errors ≤1%) were analysed.
We found suggestive evidence of an association (P≤0.05) in the SLSJ families for 29 SNPs (see Table and Tables E5, E6, E7 for the complete results). Of those, the IL1R1 rs3732131 SNP was associated with asthma in both studies with a significant association after statistical correction in the EGEA study. However, it was not fully replicated as the risk allele was not the same (pSLSJ=0.04, pEGEA=0.001, and pcombined=0.34). The association between the BACE2 rs9975388 SNP with allergic asthma was replicated in the EGEA study and reached statistical significance in the combined analysis (pSLSJ=0.05, pEGEA=0.04, and pcombined=0.004). Two SNPs in the MMP2 gene were associated with asthma in the SLSJ study (rs17301608 [pSLSJ=0.009] and rs9302671 [pSLSJ=0.01]), showed a replication in EGEA near the significance threshold (pEGEA=0.05 and pEGEA=0.07), and reached statistical significance in the combined analysis. This was observed as the same risk allele was associated in both studies (pcombined=0.002 and pcombined=0.003).
Three SNP × SNP interactions met the threshold of 5×10-4 in SLSJ and were tested for replication in EGEA (Table). All three reached statistical significance in the combined analysis after correction for multiple testing, however, none could be replicated in the EGEA study. The interaction between IL1R1*rs10208708 and IL1RAP*rs2241343 was associated with atopy (pcombined=5.4×10-5), the interaction between BACE1*rs551662 and IL1RAP*rs997534 with asthma, and the interaction between BACE1*rs676134 and IL1RAP*rs3773976 with allergic asthma (pcombined=0.01 and pcombined=0.005, respectively).
The association between IL1 and asthma has been inconsistent between studies.8 A meta-analysis revealed that there is no clear association between IL1 and asthma but shows an association the between the IL1RA (receptor antagonist) and an increased risk for the disease.8 The decoy receptor IL-1R2 has also been associated with asthma-related phenotypes34 and is involved in a decrease of inflammation in several complex diseases, such as arthritis, endometriosis, diabetes, atherosclerosis, Alzheimer's disease (AD), and ulcerative colitis.9 However, the distinct role of IL-1R2, especially in asthma and allergy remains unknown.
In this study, associations were found in the single SNP analyses for IL1R1, BACE2, MMP2, and two SNPs associated with the MMP2 gene (which was associated with asthma in an Indian population)16 survived statistical correction in the combined analysis. As IL-1R2 is a negative regulator of the IL-1 signalisation, IL-1R1 is its direct competitor for the binding of IL-1.6 The two other associated genes, BACE2 and MMP2, are known to modulate the quantity of sIL-1R2 via cleavage.617 Interestingly, sIL-1R2 seems to have a more durable interaction with IL-1β compared to the membrane-bound IL-1R2 and can also bind pro-IL-1β, being highly efficient in IL-1 sequestration.9 IL-1 is primarily involved in the initiation of the innate immune response, as well as Th17 and nonconventional Th1 cell responses. Therefore, mutations in the genes that encode its principal receptor and two enzymes that participate to the solubilisation of the decoy receptor (i.e., IL-1R2) may influence the immune response in asthma and atopy. As reported by Vasilyev and colleagues, SNPs in IL1R1 and IL1R2 modulate the level of the soluble form of these receptors on immunocompetent cells.18 Thus, it can be assumed that the associated SNPs in this study can also modulate the expression of these genes and subsequently affect the levels of IL-1R1 and IL-1R2.
The strongest gene-gene statistical interaction observed was between the IL1RAP and IL1R1 genes, and has the most biological relevance. These genes encode proteins that are associated with the regulation of the IL-1 receptor and the functionality of IL-1.6 Indeed, IL-1R1 and IL-1RAP have been shown to form a protein complex with IL-1. Furthermore, the interplay between these proteins may serve to unbalance the interaction between IL-1 and its signaling receptor (IL-1R1 and IL-1RAP) and its decoy receptor complex (IL-1R2 and IL-1RAP). This imbalance may have an important impact on the multiple roles of IL-1 in inflammation and potentially in asthma and atopy.
The interactions found between BACE1 and IL1RAP in asthma and allergic asthma is also of interest. BACE1 is known for its role in AD19 and has not been reported in relation to asthma or allergy to date. As BACE1 is involved in the inflammatory processes in AD through its modulation of IL-1B via a β-amyloid precursor protein,20 the same type of interaction may occur in asthma between BACE1 and IL-1B through IL-1RAP. Indeed, since BACE1 cleaves IL-1R217 and subsequently changes the proportion of membrane-bound and soluble forms of the receptor, the increase of sIL-1R2 may increase the competition with IL-1RAP for binding with IL-1β.
The association between the genes involved in IL-1R2 activities and asthma phenotypes have been demonstrated in this work. The power of the present study was sufficient to detect associations with the threshold of P≤0.05 for SNPs with small to moderate effects and maf ≥0.2 in the SLSJ discovery study. As the size effect of SNPs in complex traits is usually small, the power of the study surely impacts the results for the few SNPs with a lower maf (70% of the SNPs tested have maf ≥0.2) and may lead to missing some other associations between these genes and asthma or atopy. However, genes involved in IL-1R2 activity were chosen based on strong evidence of its role in asthma and asthma-related phenotypes. Moreover, the investigation of candidate genes was selected on the basis of their biological interactions with IL-1R2 and should reduce the number of false-positive findings.21
In conclusion, we suggest for the first time, that associations exist between BACE2 and MMP2 with allergic asthma and a potentially important SNP×SNP interaction between IL1RAP and IL1R1. While this genetic study does not claim to demonstrate a direct effect of these genes and SNPs on the pathophysiology of asthma, it provides a strong basis for functional studies. It will certainly achieve further fine-mapping and sequencing approaches to fully characterize the molecular structure of the genes that were associated in this study and to identify any causal variants. Our study highlights the possible role of genes modulating IL-1R2 activities in the pathophysiology of asthma, as well as asthma-related phenotypes.
Figures and Tables
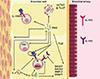 | FigureSchema of the genes involved in IL-1R2 activities analysed in this study. This figure represents the potential actions and interactions of genes involved in the IL1R2 gene activities in the bronchus of an asthmatic individual. Arrows represent the interactions between genes selected. The upper panel shows the possible involvement of the receptor IL-1R1 in the TH1 and TH17 immune response. The bottom panel illustrates the actions of IL-1R2 in the TH2 immune response. The complete information about the genes selected are available in the Supplementary Table E4. |
Table
Main association results for single SNP and SNP × SNP interaction analyses
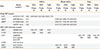
Single SNP association with P≤0.05 and SNP×SNP interaction with P≤0.0005 in SLSJ were selected for replication in the EGEA asthma study.
*Locations according to the UCSC Genome Browser assembly ID hg38.
Comb., Combination of both P values using the Stouffer's method; Freq., allele frequency of the minor allele in the SLSJ familial collection; n, number of families that contributed to the single SNP association tests and the number of individuals that contributed to the SNP×SNP interaction analysis; SNP, Single Nucleotide Polymorphism; UTR, Untranslated region.
ACKNOWLEDGMENTS
The authors thank all of the families for their valuable participation in this study. The Canada Research Chair in Genetic Determinants of Asthma, (2005-10), and the Canada Research Chair in Environment and Genetics of Respiratory Disorders and Allergy (2015-) held by C.L. enabled the maintenance and continuation of the SLSJ asthma study. C L. is the director of the Asthma Strategic Group of the Respiratory Health Network, investigator of CHILD Study and is a member of the AllerGen NCE Inc. The EGEA familial study was supported by the French National Agency for Research and GABRIEL (contract 018996 from the European Commission).
References
1. Lee SH, Park JS, Park CS. The search for genetic variants and epigenetics related to asthma. Allergy Asthma Immunol Res. 2011; 3:236–244.
2. Martinez FD, Vercelli D. Asthma. Lancet. 2013; 382:1360–1372.
3. Daley D, He JQ, Akhabir L, Stefanowicz D, Becker AB, Chan-Yeung M, et al. Associations and interactions of genetic polymorphisms in innate immunity genes with early viral infections and susceptibility to asthma and asthma-related phenotypes. J Allergy Clin Immunol. 2012; 130:1284–1293.
4. Daley D, Lemire M, Akhabir L, Chan-Yeung M, He JQ, McDonald T, et al. Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet. 2009; 125:445–459.
5. Chamberland A, Madore AM, Tremblay K, Laviolette M, Laprise C. A comparison of two sets of microarray experiments to define allergic asthma expression pattern. Exp Lung Res. 2009; 35:399–410.
6. Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013; 39:1003–1018.
7. Dinarello CA, van der Meer JW. Treating inflammation by blocking interleukin-1 in humans. Semin Immunol. 2013; 25:469–484.
8. He Y, Peng S, Xiong W, Xu Y, Liu J. Association between polymorphism of interleukin-1 beta and interleukin-1 receptor antagonist gene and asthma risk: a meta-analysis. Scientific World Journal. 2015; 685684.
9. Peters VA, Joesting JJ, Freund GG. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun. 2013; 32:1–8.
10. Colotta F, Re F, Muzio M, Bertini R, Polentarutti N, Sironi M, et al. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science. 1993; 261:472–475.
11. Laprise C. The Saguenay-Lac-Saint-Jean asthma familial collection: the genetics of asthma in a young founder population. Genes Immun. 2014; 15:247–255.
12. Kauffmann F, Dizier MH, Annesi-Maesano I, Bousquet J, Charpin D, Demenais F, et al. EGEA (Epidemiological study on the Genetics and Environment of Asthma, bronchial hyperresponsiveness and atopy)-- descriptive characteristics. Clin Exp Allergy. 1999; 29:Suppl 4. 17–21.
13. Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002; 155:478–484.
14. Gauderman WJ, Morrison JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies [Internet]. Los Angeles (CA): University of Southern California;2006. cited 2016 Jan 31. Available from: http://hydra.usc.edu/gxe.
15. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004; 74:765–769.
16. Birbian N, Singh J, Jindal SK. Highly Protective association of MMP-2-1306C/T promoter polymorphism with asthma in a North Indian population: a pilot study. Allergy Asthma Immunol Res. 2014; 6:234–241.
17. Kuhn PH, Marjaux E, Imhof A, De Strooper B, Haass C, Lichtenthaler SF. Regulated intramembrane proteolysis of the interleukin-1 receptor II by alpha-, beta-, and gamma-secretase. J Biol Chem. 2007; 282:11982–11995.
18. Vasilyev FF, Silkov AN, Sennikov SV. Relationship between interleukin-1 type 1 and 2 receptor gene polymorphisms and the expression level of membrane-bound receptors. Cell Mol Immunol. 2015; 12:222–230.
19. Cole SL, Vassar R. BACE1 structure and function in health and Alzheimer's disease. Curr Alzheimer Res. 2008; 5:100–120.
20. Gong CY, Zhou AL, Mao JH, Hu YE, Geng JS. The role of Toll-like receptor 4 on inflammation and Aβ formation in cortex astrocytes. Sheng Li Xue Bao. 2014; 66:631–638.
21. Savenije OE, Mahachie John JM, Granell R, Kerkhof M, Dijk FN, de Jongste JC, et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. J Allergy Clin Immunol. 2014; 134:170–177.
Supplementary Material
Supplementary Table E2
Power calculation for specific allele frequencies and effect sizes (odd ratios)
Supplementary Table E3
Power calculation for specific allele frequencies and effect sizes (odd ratios) for gene – gene interactions
Supplementary Table E4
Genes selected for their direct effect on IL1R2 expression or binding activity with IL-1
Supplementary Table E5
Association results for the 184 SNPs that fulfilled all quality criteria and asthma phenotype




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download