Abstract
Purpose
Drug-induced liver injury (DILI) is a serious issue often leading to discontinuation of the proper regimen of antituberculosis drugs (ATD). Previous studies have suggested that antioxidant enzymes play an important role in DILI.
Methods
We explored whether polymorphisms in superoxide dismutase genes, including Cu/Zn superoxide dismutase (SOD1), manganese superoxide dismutase (SOD2) and extracellular superoxide dismutase (SOD3) are associated with ATD-induced hepatitis. Genotype distributions of four single nucleotide polymorphisms (SNPs) in three genes (rs2070424, SOD1; rs4880, SOD2; rs2536512, and rs1799895, SOD3) were compared between 84 patients with ATD-induced hepatitis and 237 patients tolerant to ATD.
Results
Intron SNP rs2070424 of SOD1 showed a significant association with ATD-induced hepatitis. The frequency of genotypes carrying minor alleles (GA or GG) was significantly higher in the case group than that of controls (P=0.019, OR=2.26, 95% CI 1.14-4.49). For the other SNPs of SOD2 and SOD3, there were no differences in genotype frequencies between ATD-induced hepatitis and ATD-tolerant controls.
Patients in whom tuberculosis (TB) is present or strongly suspected should be treated with first-line antituberculosis drugs (ATD) including isoniazid, rifampin, pyrazinamide, and ethambutol for the first 2 months.1 This regimen is important for a curable outcome of TB; however, its use is limited by drug-induced liver injury (DILI).2 The risk of ATD-induced liver injury ranges from 5 to as high as 33%.2 In addition, the occurrence of DILI is difficult to predict. Thus, genetic markers should be used to achieve high predictability of DILI.3
While the mechanisms underlying the development of DILI remain unclear, several studies have suggested that antioxidant enzymes, such as glutathione S-transferase (GST) and superoxide dismutase (SOD), play an important role.3,4 Among these, GST acts as an intracellular scavenger by conjugating toxic reactive metabolites, which defends against oxidative stress and plays a role in detoxification.5 GSTT1 and GSTM1 null mutations are involved in the pathogenesis of ATD-induced hepatitis.6 However, our previous report did not demonstrate an association between GSTT1 and GSTM1 null mutations and ATD-induced hepatitis in Koreans.7
SOD catalyzes the dismutation of superoxide (O2-) into oxygen and hydrogen peroxide. Thus, SOD serves an important antioxidant defense against oxidative stress.8 Three forms of SOD; namely, cytoplasmic superoxide dismutase (SOD1), mitochondrial superoxide dismutase (SOD2), and extracellular superoxide dismutase (SOD3) are present in mammals.8 Two studies have reported that the SOD2 polymorphism at V16A (C47T) is significantly associated with DILI.6,9 In the present study, we hypothesized that SOD gene (SOD1, SOD2, and SOD3) polymorphisms are associated with ATD-induced hepatitis in a Korean population.
A case-control study was performed in ATD-induced hepatitis patients and ATD-tolerant patients enrolled from seven general hospitals in Korea since 2003. Diagnosis of TB in all patients was conducted in accordance with the TB guidelines.10 Criteria for inclusion included newly treated patients with first-line ATD. Patients were excluded for the following reasons: 1) An abnormal liver function test result at baseline, 2) history of liver disease (active and chronic hepatitis, fatty liver disease and cirrhosis), 3) decreased renal function, 4) other serious medical conditions requiring medication and 5) non-adherence to the treatment. All patients with pulmonary TB were treated daily with a combination regimen including INH (300-400 mg daily), RFP (450-600 mg daily), EMB (600-800 mg daily), and PZA (1,000-1,500 mg daily) for two months, and then without PZA for 4 or more months according to the treatment guideline by the American Thoracic Society, Centers for Disease Control and Prevention, and the Infectious Diseases Society of America.11 Doses of each drug were adjusted based on the body weight of the patient. Patients were monitored at regular intervals and questioned about symptoms regarding adverse reactions to ATDs. ATD-induced hepatitis was defined as an elevation in the serum levels of aspartate aminotransaminase or alanine aminotransaminase of more than three times the upper limit of the normal range during treatment and normalization of these values after cessation of treatment. ATD-tolerant controls were selected as patients with TB who did not show any ATD-related adverse reactions during the treatment period. This study was approved by the Institutional Review Boards of each participating hospital, and all subjects provided informed consent prior to participating in this study.
DNA from 43 healthy volunteers was sequenced for SOD gene polymorphisms using an ABI Prism 3100 DNA analyzer (Applied Biosystems, Forster city, CA, USA). We then tagged SNPs for genotyping among the informative SNPs with minor allele frequencies greater than 0.02 and r2 greater than 0.8. Four polymorphisms were selected in the SOD genes; 1 in SOD1 (Ivs3-251A/G), 1 in SOD2 (V16A) and 2 in SOD3 (H63R and A236G). The minor allele frequencies of Ivs3-251A/G, V16A, H63R, and A236G were 0.491, 0.109, 0.299, and 0.027, respectively.
Genomic DNA was prepared from peripheral blood samples using the Genomic PUREGENE® DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, MN, USA). Polymorphisms in SOD genes were genotyped using the SNaPshot assay according to the manufacturer's instructions (ABI PRISM SNaPShot Multiplex kit, Foster City, CA, USA). Analysis was performed using Genemapper software (version 4.0; Applied Biosystems). The primer sets and Tm used for the SNaPshot assay are provided in Table 1.
Genotype frequencies were compared between the 2 groups and examined using a multivariate logistic regression analysis controlling for age and gender as covariates. Hardy-Weinberg equilibrium was examined using chi-square tests. All statistical analyses were performed using SAS (version 8.1; SAS Institute, Cary, NC, USA), and P values of less than 0.05 were considered to indicate statistical significance.
A total of 84 patients with ATD-induced hepatitis and 237 patients with ATD-tolerance were enrolled. Of the 84 enrolled subjects with ATD-induced hepatitis, skin rash was present in 15 (17.8%) patients. All subjects were ethnically Korean. The number of females was significantly higher in patients with ATD-induced hepatitis than controls (48.8% vs 31.6%, respectively; P<0.01). There were no significant differences in demographic parameters such as age, height, weight, body mass index, and basal AST and ALT levels between the case and control subjects (Table 2).
Genotype frequencies of the four polymorphisms in three SOD genes; SOD1, SOD2, and SOD3, were analyzed and evaluated for an association with the development of ATD-induced hepatitis (Table 3). In SOD1, IVS3-251A/G polymorphism in introns was significantly associated with ATD-induced hepatitis with higher frequencies of genotypes carrying a minor allele (AG or GG) in the case group compared to controls (85.7% vs 72.5%, P=0.019, OR=2.26, 95%, CI: 1.14-4.49). The genotype frequencies of SOD2 SNP (V16A) showed no differences between case and control subjects. Neither of the two selected SOD3 SNPs (H63R and A236G) was significantly associated with the development of ATD-induced hepatitis based on a logistic regression analysis.
This study showed a significant association between IVS3-251A/G SOD1 polymorphisms in intron and ATD-induced hepatitis in a Korean population. In addition, there were no differences in genotype frequencies of others SNPs, including SOD2 V16A, which is known to be associated with DILI in Taiwanese and Spanish populations.6,9
Reactive oxygen species (ROS) overproduction in response to various toxins and the following cellular damage is commonly implicated in DILI pathogenesis.4 CYP450 enzymes, GST, and SOD are essential for antioxidant defense; therefore, they should be considered candidate genes associated with DILI development.3 Our previous pharmacogenetic study suggested that genetic variants in N-acetyltransferase 2 (NAT2) may increase the risk of anti-tuberculosis drug-induced hepatitis.12 However, there was no significant association between genotypes of CYP2C9, CYP2C19, CYP2D6, CYP2E1, GSTT1, and GSTM1 and ATD-induced hepatitis.7,12
The SOD1 gene has been localized to chromosome 21 (region 21q22) in humans and contains four introns and five exons. SOD1 knock-out mice are known to be resistant to acetaminophen toxicity.8,13 This is the first report to identify an association between IVS3-251A/G polymorphisms in the SOD1 gene and DILI. Previously, IVS3-251A/G polymorphisms in the SOD1 gene were associated only with myelomenigocele and sensorineural hearing loss.14,15 However, the functional significance of this polymorphism in SOD1 remains unclear. Previous studies reported that rare alleles of the four SNPs (rs202446, rs202447, rs4816405, and rs2070424) in SOD1 are associated with an increased expression of SOD1 mRNA in the CEPH cell lines.15 On the contrary, Liu et al.14 reported that plasma activity of SOD1 enzyme, total anti-oxidation capability (T-AOC), and plasma malondialdehyde (MDA) content was not significantly associated with the genotypes of SNP rs2070424. Additional functional studies are required to explore the role of IVS3-251A/G polymorphisms in SOD1 in the pathogenesis of DILI.
Our study did not identify an association between SOD2 V16A and ATD-induced hepatitis in a Korean population. This result is not in agreement with a previous study by Huang and coworkers,6 who reported that the SOD2 Ala/Ala and Ala/Val genotypes increased the risk of developing DILI in the Taiwanese population. In addition, Lucena et al.9 reported that the SOD2 V16A polymorphism is associated with DILI in the Spanish population. However, our results differed from the Taiwanese data, which showed that carriers of the SOD2 Ala/Ala genotype were at higher risks of developing cholestatic and mixed types of DILI, but not hepatocellular-type, such as ATD-induced hepatitis.9 There are several reasons for these discrepancies. First, ethnic differences in allele frequencies can explain the variability in genetic studies related to DILI.16 SOD2 genotype distribution in Koreans (TT/TC/CC:78.9/20.2/0.8%) differed between the Spanish (31/49/20%) and Taiwanese (75.7/21.7/2.6%) controls, which could have an impact on the expression of liver injury.6,9 Second, a wide range of drugs are known to induce DILI and the diverse characteristics of these drugs are involved in DILI pathogenesis. Furthermore, genetic susceptibility to DILI can be drug-specific.16 All DILI cases in our cohort were ATD-induced DILI, whereas 55% of the Taiwanese cohort and only 3% of the Spanish cohort showed ATD-induced DILI.6,9
The lack of functional characterization is the major limitation of our study. Therefore, we could not determine the precise mechanisms underlying associations between genotypes and phenotypes. Second, we enrolled patients with ATD-induced hepatitis without identifying the responsible drug, which can mimic genetic risk factors in SOD genes and can be associated with ATD-induced hepatitis. Third, we evaluated only one SNP in SOD1 or SOD2. Although the frequencies of other SNPs in SOD1 or SOD2 were less than 2%, they may be positively associated with ATD-induced hepatitis.
In conclusion, intron SNP Ivs3-251A/G in SOD1 (rs2070424) showed a significant association with ATD-induced hepatitis. There was no difference in genotype frequencies of others SNPs of SOD2 and SOD3, including SOD2 V16A, which showed a significant association with DILI in Taiwan. Some data comparing ethnicities suggests that it is difficult to identify a global genetic marker for ATD-induced liver injury. However, further studies should explore these markers to achieve high predictability for ATD-induced liver injury in a Korean population.
Figures and Tables
Table 1
Primer sets and Tm used for the SNaPshot assay

Table 2
Clinical characteristics of the study subjects
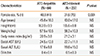
Table 3
Genotype frequencies of SOD1, SOD2, and SOD3 polymorphisms in patients with ATD-induced hepatitis and in the ATD-tolerant control
ACKNOWLEDGMENTS
This study was supported by a grant of the National Project for Personalized Genomic Medicine, Ministry for Health & Welfare, Republic of Korea (A111218-11-PG01).
References
1. American Thoracic Society. CDC. Infectious Diseases Society of America. Treatment of tuberculosis. MMWR Recomm Rep. 2003; 52:1–77.
2. Saukkonen JJ, Cohn DL, Jasmer RM, Schenker S, Jereb JA, Nolan CM, Peloquin CA, Gordin FM, Nunes D, Strader DB, Bernardo J, Venkataramanan R, Sterling TR. ATS (American Thoracic Society) Hepatotoxicity of Antituberculosis Therapy Subcommittee. An official ATS statement: hepatotoxicity of antituberculosis therapy. Am J Respir Crit Care Med. 2006; 174:935–952.
3. Russmann S, Jetter A, Kullak-Ublick GA. Pharmacogenetics of drug-induced liver injury. Hepatology. 2010; 52:748–761.
4. Kaplowitz N. Drug-induced liver injury. Clin Infect Dis. 2004; 38:Suppl 2. S44–S48.
5. Huang YS. Genetic polymorphisms of drug-metabolizing enzymes and the susceptibility to antituberculosis drug-induced liver injury. Expert Opin Drug Metab Toxicol. 2007; 3:1–8.
6. Huang YS, Su WJ, Huang YH, Chen CY, Chang FY, Lin HC, Lee SD. Genetic polymorphisms of manganese superoxide dismutase, NAD(P)H:quinone oxidoreductase, glutathione S-transferase M1 and T1, and the susceptibility to drug-induced liver injury. J Hepatol. 2007; 47:128–134.
7. Kim SH, Kim SH, Yoon HJ, Shin DH, Park SS, Kim YS, Park JS, Jee YK. GSTT1 and GSTM1 null mutations and adverse reactions induced by antituberculosis drugs in Koreans. Tuberculosis (Edinb). 2010; 90:39–43.
8. Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med. 2009; 47:344–356.
9. Lucena MI, García-Martín E, Andrade RJ, Martínez C, Stephens C, Ruiz JD, Ulzurrun E, Fernandez MC, Romero-Gomez M, Castiella A, Planas R, Durán JA, De Dios AM, Guarner C, Soriano G, Borraz Y, Agundez JA. Mitochondrial superoxide dismutase and glutathione peroxidase in idiosyncratic drug-induced liver injury. Hepatology. 2010; 52:303–312.
10. World Health Organization. Treatment of tuberculosis: guidelines. 4th ed. Geneva: World Health Organization;2010.
11. Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, Fujiwara P, Grzemska M, Hopewell PC, Iseman MD, Jasmer RM, Koppaka V, Menzies RI, O'Brien RJ, Reves RR, Reichman LB, Simone PM, Starke JR, Vernon AA. American Thoracic Society, Centers for Disease Control and Prevention and the Infectious Diseases Society. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med. 2003; 167:603–662.
12. Kim SH, Kim SH, Bahn JW, Kim YK, Chang YS, Shin ES, Kim YS, Park JS, Kim BH, Jang IJ, Song J, Kim SH, Park HS, Min KU, Jee YK. Genetic polymorphisms of drug-metabolizing enzymes and anti-TB drug-induced hepatitis. Pharmacogenomics. 2009; 10:1767–1779.
13. Lei XG, Zhu JH, McClung JP, Aregullin M, Roneker CA. Mice deficient in Cu,Zn-superoxide dismutase are resistant to acetaminophen toxicity. Biochem J. 2006; 399:455–461.
14. Liu YM, Li XD, Guo X, Liu B, Lin AH, Rao SQ. Association between polymorphisms in SOD1 and noise-induced hearing loss in Chinese workers. Acta Otolaryngol. 2010; 130:477–486.
15. Kase BA, Northrup H, Morrison AC, Davidson CM, Goiffon AM, Fletcher JM, Ostermaier KK, Tyerman GH, Au KS. Association of copper-zinc superoxide dismutase (SOD1) and manganese superoxide dismutase (SOD2) genes with nonsyndromic myelomeningocele. Birth Defects Res A Clin Mol Teratol. 2012; 94:762–769.
16. Andrade RJ, Agúndez JA, Lucena MI, Martínez C, Cueto R, García-Martín E. Pharmacogenomics in drug induced liver injury. Curr Drug Metab. 2009; 10:956–970.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download