Abstract
Purpose
We investigated maternal genetic effects of four IL-4/IL-13 pathway genes as well as their interactions with the "Western or Eastern lifestyles/environments" on IgE in Karelian children.
Methods
This study included 609 children and their mothers. Total IgE levels in children and mothers were measured and 10 single nucleotide polymorphisms (SNPs) in IL-4, IL-4Ra, IL-13, and STAT6 were genotyped in mothers and their children.
Results
The maternal G allele of IL-13 130 (rs20541) was significantly (P=0.001) associated with decreased IgE in children in the Karelian population (Pooling Finnish and Russian children), as well as in Finnish (P=0.030) and Russian children (P=0.018). The IgE levels were significantly (P=0.001) higher in Russian children whose mothers were homozygous for the G allele of the IL-4Ra 50 (rs1805010) SNP than that in Russian children of mothers who were AG heterozygotes or AA homozygotes. After accounting for children's genotypes, we observed interactive effects on children's IgE for maternal IL-13 130 genotypes (P=0.014) and maternal IL-4Ra 50 genotypes (P=0.0003) with "Western or Eastern" lifestyles/environments. With the adjustment for multiple comparisons using a false discovery rate (FDR) of 0.05, the interactive effect of the maternal IL-4Ra50 SNP was significant.
Immunoglobulin E (IgE) is a monomeric antibody that has been implicated in immune responses to helminth infections.1 High total serum IgE levels are a clinical characteristic of asthma and atopy in western communities2 and have also been associated with a number of environmental and genetic risk factors.3 Twin registry studies have consistently shown a significant genetic component to serum IgE variation across individuals in all age groups, with cord blood, infants, children and adults levels having heritability ranging from 30% to 70%.4,5 Candidate gene associated studies have also shown that genetic variants in a number of genes, particularly in interleukin (IL)-4/IL-13 signal pathway genes, were associated with IgE levels.6,7,8
IL-13 and IL-4, which are essential for IgE synthesis, share a common signalling pathway in binding to a heterodimer of the IL-4 receptor α and either the common γ-chain (for binding IL-4 and IL-13) or the IL-13 receptor α chain (exclusive for IL-13).9 By phosphorylation of the transcriptional regulator of signal transducer and activator of transcription 6 (STAT6), intracellular signalling of IL4/IL-13 pathway penetrates the nucleus and activates the transcription of target genes inducing IgG switching to IgE.10 Several functionally important genetic variants in the IL-4/IL-13 pathway genes should have an influence on serum IgE levels in view of the critical role of the IL-4/IL-13 pathway in promotion of production of IgE. We and others have previously reported the associations of those genetic variants with IgE.11,12,13
Maternal genetic variants may determine the prenatal environment and thus play a role in the development of health conditions in their offspring, particularly for diseases with onset early in life.14,15 Many prenatal factors have been associated with IgE levels later in life.16,17 It is, therefore, conceivable that maternal genetic variants may have an influence on IgE levels in school-age children, particularly for genes in the IL-4/IL-13 pathway, as there is evidence that cord blood IgE was a predictor for allergy and asthma later in life.18,19 However, no reported studies have investigated the effects of maternal genetic variants in IL4/IL13 pathway genes on IgE levels in children. The Karelian population provides a unique opportunity to clarify the maternal genetic effects on IgE in school-age children. In the present study, we hypothesised that maternal genetic variants in 4 IL-4/IL-13 pathway genes would have an influence on IgE levels in Karelian school-age children. As offspring share half their genes with their mothers, it is challenging to differentiate between conditions due to the influence of maternal genetic effects on fetal development and those due to the offspring's inherited genetic effects.20 In the present study, we investigated the maternal genetic effects on serum total IgE in children conditioning on the corresponding offspring's genotypes for 4 IL-4/IL-13 pathway genes.
Karelian children living on the Finnish border had significantly higher prevalence of asthma and allergy compared to those living on the Russian border.21 Our previous studies have shown that there are significant interactive effects on asthma-related phenotypes between genetic variants in several asthma candidate genes and "Western or Eastern lifestyles/environments" in the Karelian population.11,22 The secondary hypothesis, for the present study, is that if the maternal genetic effects on the 4 IL-4/IL-13 pathway genes influence IgE levels in children these factors would interact with the "Western or Eastern lifestyles/environments" in the Karelian population.
The Karelian population has been described previously.23,24 Briefly, the Karelian children and their mothers were recruited through 24 schools in Finland and 11 schools in Russia in 2003. The Finnish study area, North Karelia, is the most eastern province of Finland and the Russian study area is on the other side of the border, which was part of the Soviet Union until the collapse of the country in 1991. The Finnish and Russian Karelian populations are ethnically mainly European, and it is assumed that they are a genetically homogeneous population that experienced different "Western" and "Eastern" environments/lifestyles after World War Two.
The present study included 609 children and their mothers (334 Finnish and 275 Russian). The study was approved by the Ethics Committee of the Helsinki University Central Hospital.Potential confounding variables related to levels of IgE and allergy such as passive smoking, farm exposure in the first year of life, pet exposure, number of siblings and other socio-economic factors have been comprehensively investigated, and symptoms of allergy and asthma have been assessed, in the study population. Total serum IgE levels in children and mothers were measured with the Pharmacia CAP System (Pharmacia Upjohn, Uppsala, Sweden).
Genomic DNA was extracted by an automated DNA extraction instrument (Autopure LS; Qiagen, Hilden, Germany). Ten single nucleotide polymorphisms (SNPs) in IL-4, IL-4Ra, IL-13, and STAT6 were selected and genotyped based on their functional importance and evidence of associations with asthma and IgE.25 Genotyping of candidate polymorphisms was performed using iPLEX™ Assay on the MassARRAY® system (Sequenom, San Diego, CA, USA) according to manufacturer's instructions (http://www.sequenom.com). All genotyping was conducted by the Australian Genome Research Facility.
Exact Hardy-Weinberg Equilibrium (HWE) testing was conducted to examine the HWE violation of each polymorphism in Finnish and Russian children and their mothers, separately. To investigate the association of maternal and offspring's genotypes with total serum IgE levels, analysis of variance (ANOVA), independent sample t-test where appropriate, was utilised. To clarify the maternal effects of the investigated genotypes, children were stratified by their own genotypes of the corresponding SNP. The genetic effects of these polymorphisms were analysed in the 2 populations separately as well as in the whole population (pooling Finnish and Russian children together). The interactive effects were investigated using the Likelihood-ratio test after estimation in a linear regression in STATA. Both the unrestricted (with interaction) and the restricted (without interaction) models were fitted using the maximum likelihood method and the 2 models were compared to identify the significant interactions. To adjust for potential confounders, multivariate regression analyses were employed. To accounting for multiple hypothesis testing we employed false discovery rate (FDR) control with a FDR of 0.05.
The 10 SNPs were in Hardy-Weinberg Equilibrium in Finnish and Russian mothers (Table 1). There were significant differences in genotype frequencies of IL-4 -589, IL-4 -33, IL-4 2979, IL-13 130, and IL-13 4738 between Finnish and Russian mothers. The SNPs of IL-4 -589 and IL-13 4738 were in complete linkage disequilibrium with the SNPs of IL-4 -33 in the IL-4 gene and IL-13 130 in the IL-13 gene, respectively, therefore only the results of IL-4 -33 and IL-13 130 were reported.
The geometric mean of total IgE in Finnish children was 107.3 (95%CI: 90.4-127.4) kU/L and was not significantly higher than the mean of 91.3 (79.7-104.6) kU/L in Russian children. Finnish mothers had significantly (P=0.005) lower levels of total IgE (42.3 [95% CI: 36.7-48.7] kU/L) compared to Russian mothers (56.6 [49.0-65.3] kU/L). The correlation coefficients of total IgE levels in children and their mothers were 0.18 (P=0.001) and 0.39 (P<0.001) in Finnish and Russian Karelians, respectively.
We investigated the association of maternal genotypes of the 8 SNPs with maternal serum total IgE in the Karelian population (pooling Finnish and Russian Karelians together) as well as in Finnish and Russian mothers, separately. No association was found in the pooled Karelian population between maternal genotype and maternal IgE. In the split populations only the T allele of IL-4Ra 478 (P=0.063) and the A allele of IL-4Ra 551 (P=0.073) were insignificantly associated with increased levels of maternal IgE in Russian mothers.
We examined the association of maternal genotypes with children' serum total IgE. The G allele of IL-13 130 was significantly (P=0.001 for linear trend and between group [ANOVA]) associated with decreased IgE in the Karelian population (as a whole, Fig. 1A), as well as in Finnish (P=0.030 for linear trend and P=0.006 for between groups [ANOVA], Fig. 1B) and Russian children (P=0.018 for linear trend and P=0.039 for between groups [ANOVA], Fig. 1C). In the whole population, the SNP of IL-4Ra 50 was not associated with IgE levels in children (Fig. 2A). However, in Russian Karelians the maternal variants of SNP of IL-4Ra 50 was associated with IgE in children (Fig. 2C). In 47 Russian children whose mothers were homozygous for the G allele of the IL-4Ra 50 SNP the serum IgE levels (mean, 95% CIs: 153.3, 109.4-214.0 kU/L) were significantly (P=0.001) higher than that (76.2, 63.8-91.2 kU/L) in Russian Karelian children (n=220) of mothers who were AG heterozygotes or AA homozygotes. In Finnish children maternal GG genotypes (homozygous for the G allele of the IL-4Ra 50 SNP) were not associated with increased IgE (Fig. 2B). We did not find any other significant associations of maternal genetic variants in the 8 SNPs of IL-4/IL-13 pathway genes with children's serum IgE in the whole and stratified populations.
We have previously reported that the G allele of IL-13 130 in children was significantly associated with decreased children's IgE in the Karelian population.11 The above mentioned maternal genetic effects on children's IgE may partly be attributable to shared genetic codes between mothers and their children. To clarify the maternal genetic effects, we investigated these associations for maternal genotypes stratified by the child's genotypes. Finnish children homozygous for the G allele of IL-13 130 had significantly (P=0.01, Fig. 3A) lower serum IgE (mean: 49.0; 95% CI: 31.9-74.9 kU/L) levels if their mothers were also GG homozygous (n=58) compared to those whose mothers were AG heterozygotes (n=47; mean: 111.6; 95% CI: 69.8-178.1 kU/L). The maternal effect of the G allele did not exist (P=0.62, Fig. 3B) in Russian children homozygous for the G allele and with mothers being GG (n=88; mean: 69.4; 95% CI: 52.3-91.9 kU/L) compared to those whose mothers were AG (n=49; mean: 61.7; 95% CI: 42.3-89.7 kU/L).
Moreover, for the IL-4Ra 50 SNP, considering heterozygous AG Finnish children (Fig. 4), the maternal G allele was significantly (P=0.032 for linear trend) associated with decreased serum IgE in Finnish children with means of 63.4, 105.8, and 155.3 kU/L for maternal GG, AG and AA genotypes, respectively. In the heterozygous AG Russian children, the maternal GG genotype appeared to be associated with the significantly (P=0.00006) highest serum IgE levels in their children (n=22; mean: 220.5; 95% CI: 142.8-340.6 kU/L) relative to maternal AG (n=61; mean: 58.7; 95% CI: 43.4-79.4 kU/L) and AA genotypes (n=31; mean: 68.9; 95% CI: 43.2-110.1 kU/L) (Fig. 4B).
We examined the interactive terms of maternal genotypes and the variable representing "Western or Eastern" lifestyles/environments after stratifying by the children's genotypes for the 8 SNPs (Table 2). We found that the interactive effect on children's IgE was significant (P=0.014) for maternal IL-13 130 genotypes and "Western or Eastern" lifestyles/environments in children who were homozygous for the G allele of this SNP. Likewise, the interactive effects were significant (P=0.0003) for maternal IL-4Ra 50 genotypes and "Western or Eastern" lifestyles/environments in children who were heterozygous AG of the IL-4Ra 50 SNP. Several confounding variables including age, gender, number of children at home, parents' education, residential suburb housing, passive smoking, farm exposure in the first year of life, farm animal exposure, cat and dog exposure were investigated for associations with serum IgE levels in children. Only gender and dog exposure had an effect on IgE levels with a significance of P<0.10. The 2 confounding variables were added in the model when the interactive effects of maternal genotypes were investigated. After we adjusted for these two confounders the significance of the interactive effects of the 2 maternal SNPs reminded (Table 2). Using FDR control techniques, the interactive effect of the maternal IL-4Ra 50 SNP on IgE in children who were heterozygous AG of the IL-4Ra 50 SNP was still significant (the boundary significance is 0.002 with a FDR of 0.05).
We investigated the effects of maternal genetic variants in the four IL-4/IL-13 pathway genes, IL-4, IL-4Ra, IL-13, and STAT6, on school children's serum total IgE. Consistent with our hypothesis, we confirmed that maternal genetic variants had an influence on children's IgE. The effects of the SNP IL-4Ra 50 on children's IgE were significant after correcting for the multiple tests using FDR control techniques. Although the effects of maternal IL-13 130 were not significant with the correction, the finding of maternal genetic effects for this SNP was still important and warrants further investigations, recognizing the functional importance of the IL-13 gene and the consistent associations between the SNP and IgE levels previously reported.25,26,27 This study is the first to have identified the maternal genetic effects of the IL-4/IL-13 pathway genes on school-age children's serum IgE. We also found that the maternal genetic effects of the IL-4Ra 50 and IL-13 130 SNPs differed in Finnish and Russian Karelian populations. This indicates that the contrasting environments/lifestyles in Finnish and Russian Karelians may have modified the maternal genetic effects in relation to the production of IgE as well as IgE-related phenotypes in children. These findings in this study have significantly improved our understanding of the mechanisms regarding maternal genetic effects of IL-4/IL-13 pathway genes and gene-environment interactions that underlie the development of IgE-related phenotypes in children.
The maternal genetic predisposition can have a significant influence on the offspring's phenotypes as a provider of genes. However, it is important to clarify whether the maternal genetic variants have effects on their children's phenotypes, such as asthma and allergy, which are independent of the offspring's inherited maternal genetic variants. In the present study, we stratified for the children's genotypes and examined the maternal genetic effects of the corresponding SNPs. We confirmed that some genetic variants in the IL-4/IL-13 pathway genes have independent maternal genetic effects on children's IgE. There are several plausible explanations for the maternal genetic effects. Firstly, the maternal genetic variants may influence the offspring's disease susceptibility through phenotypes expressed during pregnancy. It is generally agreed that many aspects of health and disease are determined prenatally. Consistent evidence has shown that prenatal risk factors have influences on IgE, asthma and allergy. Cord blood IgE levels have been reported to be associated with the development of asthma and allergy later in life.18,19 Maternal genetic variants in IL-4/IL-13 pathway genes may have influenced fetal blood IgE or T helper cell 1(Th1)/Th2 balance during pregnancy, thereby leading to their offspring having a high production of IgE. Secondly, apart from the in utero environment, many other factors that may have significant influences on the development of health and disease in infants and children are also under the control of the maternal genome. For example, breast feeding and pre/antenatal environmental exposures, maternal behaviour and socio-economic status, may interact with maternal genetic effects. Thirdly, the maternal genetic effects may partly be attributable to epigenetics. Consistently, maternal history of allergic disease is a stronger predictor for offspring's susceptibility compared to paternal history.28 This evidence and our findings all point to epigenetic influences29 that warrant more studies. In addition, considering the possible complex relationships between maternal genetics and children's IgE further studies are necessary to clarify the maternal genetic effects of IL4/IL13 pathway genes.
We have previously reported that the prevalences of asthma and allergy were significantly higher in Finnish Karelians compared to Russian Karelians. However, the total IgE levels were higher in Russian Karelian mothers than Finnish Karelian mothers and the difference in total IgE levels for school-age children was not significant. The high levels of total IgE in Russian Karelians may be due to the potential high prevalence of helminth infections in the Russian community.30 However, it was reported that the levels of specific IgE to common environmental allergens were significant higher in Karelian Finnish children than Russian children, although there was a significant disparity in the association of asthma, rhinitis, and eczema with allergen-specific IgE between Finnish and Russian Karelians.23
The present study found that the maternal genetic effects on children's total IgE levels varied in Finnish and Russian Karelians. We have previously reported that genetic variants in several asthma candidate genes in mothers and children had opposite effects on asthma and allergy in Finnish and Russian Karelians. The findings in this study further exemplify the complex gene-environment interactions,31,32 for example with maternal genetic effects and their offspring's phenotypes. These findings may partly explain the significant gradients of asthma and allergic conditions between Western and Eastern environment/lifestyles, and this aspect requires further epigenetic studies.
We did not find significant associations between maternal genetic variants in the IL-4/IL13 pathway genes and mothers' serum IgE, even for the SNP IL-13 130 that has been consistently associated with IgE levels. The life-time environmental exposures of the adult women may have distorted the genetic effects, highlighting the importance of gene-environment interactions in the development of IgE-related phenotypes such as asthma and allergy.
One limitation to the present study was that sample sizes were limited when we investigated the maternal genetic effects after stratifying by the Finnish and Russian children's genotypes. However, despite this sample size we have identified a significant maternal genetic effect as well as interactions with "Western or Eastern" environments/lifestyles after correcting for multiple tests. Levels of IgE are a continuous variable that as an outcome has a relatively larger statistical power for detecting a genotype-phonotype association than a categorical variable. Although there were 609 children and their mothers in the study, we acknowledge it lacked power to investigate genotype and phenotype relationships for categorical variables such as asthma and rhinitis, and gene-environment interactions, particularly for maternal genetic effects, after stratifying for the children's genotypes. Consequently, the results should be interpreted with caution and we encourage further studies to confirm these findings.
In conclusion, maternal genetic variants in IL-4/IL-13 pathway genes, such as IL-13 130 and IL-4Ra 50, influenced IgE levels in school-age children that were independent of the children's genetic effects. These maternal genetic effects were interacting with "Western or Eastern" environments/lifestyles, thereby determining the capacity of IgE production in children.
Figures and Tables
Fig. 1
Serum Total IgE in children and maternal genotypes of IL-13 130. (A) Karelian (Finnish+Russian), (B) Finnish, (C) Russian.
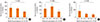
Fig. 2
Serum Total IgE in children and maternal genotypes of IL-4RA 50. (A) Karelian (Finnish+Russian), (B) Finnish, (C) Russian.
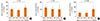
Fig. 3
The associations of maternal genotypes of IL-13 130 and IgE in children homozygous for the G allele of IL-13 130 (n=242). (A) Finnish, (B) Russian.

Fig. 4
The associations of maternal genotypes of IL-4RA 50 and IgE in children heterozygous AG of IL-4RA 50 (n=254). (A) Finnish, (B) Russian.
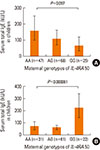
Table 1
Frequencies of the 10 polymorphisms in the four IL-4/IL-13 pathway genes in Finnish and Russian Karelians (Mothers)
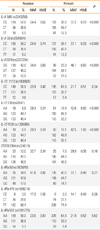
Table 2
The interactive effects of maternal genotypes with the "Western or Eastern lifestyles/environments" on serum total IgE in children stratified by the corresponding offspring's genotypes
ACKNOWLEDGMENTS
This work was supported by the European Commission's Seventh Framework Programme under grant agreement No. 261357. GZ is supported by NHMRC and BrightSpark Foundation.
References
1. Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003; 21:579–628.
2. Broide DH, Finkelman F, Bochner BS, Rothenberg ME. Advances in mechanisms of asthma, allergy, and immunology in 2010. J Allergy Clin Immunol. 2011; 127:689–695.
3. Wang TN, Tseng HI, Kao CC, Chu YT, Chen WY, Wu PF, Lee CH, Ko YC. The effects of NOS1 gene on asthma and total IgE levels in Taiwanese children, and the interactions with environmental factors. Pediatr Allergy Immunol. 2010; 21:1064–1071.
4. Hanson B, McGue M, Roitman-Johnson B, Segal NL, Bouchard TJ Jr, Blumenthal MN. Atopic disease and immunoglobulin E in twins reared apart and together. Am J Hum Genet. 1991; 48:873–879.
5. Jacobsen HP, Herskind AM, Nielsen BW, Husby S. IgE in unselected like-sexed monozygotic and dizygotic twins at birth and at 6 to 9 years of age: high but dissimilar genetic influence on IgE levels. J Allergy Clin Immunol. 2001; 107:659–663.
6. Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006; 117:269–274.
7. Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, Weiland SK, Erickson RP, von Mutius E, Martinez FD. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000; 105:506–513.
8. Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin JM. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998; 19:119–120.
9. LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008; 132:259–272.
10. Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999; 17:701–738.
11. Zhang G, Candelaria P, Mäkelä JM, Khoo SK, Hayden MC, von Hertzen L, Laatikainen T, Vartiainen E, Goldblatt J, Haahtela T, LeSouef NP. Disparity of innate immunity-related gene effects on asthma and allergy on Karelia. Pediatr Allergy Immunol. 2011; 22:621–630.
12. Hunninghake GM, Soto-Quirós ME, Avila L, Su J, Murphy A, Demeo DL, Ly NP, Liang C, Sylvia JS, Klanderman BJ, Lange C, Raby BA, Silverman EK, Celedón JC. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007; 120:84–90.
13. Yabiku K, Hayashi M, Komiya I, Yamada T, Kinjo Y, Ohshiro Y, Kouki T, Takasu N. Polymorphisms of interleukin (IL)-4 receptor alpha and signal transducer and activator of transcription-6 (Stat6) are associated with increased IL-4Ralpha-Stat6 signalling in lymphocytes and elevated serum IgE in patients with Graves' disease. Clin Exp Immunol. 2007; 148:425–431.
14. Romero R, Friel LA, Velez Edwards DR, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, Pearce BD, Bartlett J, Salisbury BA, Anant MK, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Williams SM, Menon R. A genetic association study of maternal and fetal candidate genes that predispose to preterm prelabor rupture of membranes (PROM). Am J Obstet Gynecol. 2010; 203:361e1–e30.
15. Swamy GK, Garrett ME, Miranda ML, Ashley-Koch AE. Maternal vitamin D receptor genetic variation contributes to infant birthweight among black mothers. Am J Med Genet A. 2011; 155A:1264–1271.
16. Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, Schram-Bijkerk D, Brunekreef B, van Hage M, Scheynius A, Pershagen G, Benz MR, Lauener R, von Mutius E, Braun-Fahrländer C. Parsifal Study Team. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006; 117:817–823.
17. Miller RL. Prenatal maternal diet affects asthma risk in offspring. J Clin Invest. 2008; 118:3265–3268.
18. Pesonen M, Kallio MJ, Siimes MA, Elg P, Björksten F, Ranki A. Cord serum immunoglobulin E as a risk factor for allergic symptoms and sensitization in children and young adults. Pediatr Allergy Immunol. 2009; 20:12–18.
19. Wen HJ, Wang YJ, Lin YC, Chang CC, Shieh CC, Lung FW, Guo YL. Prediction of atopic dermatitis in 2-yr-old children by cord blood IgE, genetic polymorphisms in cytokine genes, and maternal mentality during pregnancy. Pediatr Allergy Immunol. 2011; 22:695–703.
20. Mitchell LE, Weinberg CR. Evaluation of offspring and maternal genetic effects on disease risk using a family-based approach: the "pent" design. Am J Epidemiol. 2005; 162:676–685.
21. Vartiainen E, Petäys T, Haahtela T, Jousilahti P, Pekkanen J. Allergic diseases, skin prick test responses, and IgE levels in North Karelia, Finland, and the Republic of Karelia, Russia. J Allergy Clin Immunol. 2002; 109:643–648.
22. Zhang G, Khoo SK, Laatikainen T, Pekkarinen P, Vartiainen E, von Hertzen L, Hayden CM, Goldblatt J, Mäkelä M, Haahtela T, Le Souëf PN. Opposite gene by environment interactions in Karelia for CD14 and CC16 single nucleotide polymorphisms and allergy. Allergy. 2009; 64:1333–1341.
23. Pekkarinen PT, von Hertzen L, Laatikainen T, Mäkelä MJ, Jousilahti P, Kosunen TU, Pantelejev V, Vartiainen E, Haahtela T. A disparity in the association of asthma, rhinitis, and eczema with allergen-specific IgE between Finnish and Russian Karelia. Allergy. 2007; 62:281–287.
24. von Hertzen L, Mäkelä MJ, Petäys T, Jousilahti P, Kosunen TU, Laatikainen T, Vartiainen E, Haahtela T. Growing disparities in atopy between the Finns and the Russians: a comparison of 2 generations. J Allergy Clin Immunol. 2006; 117:151–157.
25. Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008; 8:169–182.
26. Leung TF, Tang NL, Chan IH, Li AM, Ha G, Lam CW. A polymorphism in the coding region of interleukin-13 gene is associated with atopy but not asthma in Chinese children. Clin Exp Allergy. 2001; 31:1515–1521.
27. Wang M, Xing ZM, Lu C, Ma YX, Yu DL, Yan Z, Wang SW, Yu LS. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum Genet. 2003; 113:387–390.
28. Litonjua AA, Carey VJ, Burge HA, Weiss ST, Gold DR. Parental history and the risk for childhood asthma. Does mother confer more risk than father? Am J Respir Crit Care Med. 1998; 158:176–181.
29. Fedulov AV, Kobzik L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol. 2011; 44:285–292.
30. Matthys B, Bobieva M, Karimova G, Mengliboeva Z, Jean-Richard V, Hoimnazarova M, Kurbonova M, Lohourignon LK, Utzinger J, Wyss K. Prevalence and risk factors of helminths and intestinal protozoa infections among children from primary schools in western Tajikistan. Parasit Vectors. 2011; 4:195.
31. Zhang G, Goldblatt J, LeSouëf P. The era of genome-wide association studies: opportunities and challenges for asthma genetics. J Hum Genet. 2009; 54:624–628.
32. Zhang G, Goldblatt J, LeSouëf PN. Does the relationship between IgE and the CD14 gene depend on ethnicity? Allergy. 2008; 63:1411–1417.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download