Abstract
Purpose
Cockroach (CR) is a common source of indoor allergens, and Per a 1 is a major American CR (Periplaneta americana) allergen; however, several attributes of this protein remain unknown. This study identifies a novel specific B cell epitope and anatomical locations of Per a 1.0105.
Methods
Recombinant Per a 1.0105 (rPer a 1.0105) was used as BALB/c mouse immunogen for the production of monoclonal antibodies (MAb). The MAb specific B cell epitope was identified by determining phage mimotopic peptides and pair-wise alignment of the peptides with the rPer a 1.0105 amino acid sequence. Locations of the Per a 1.0105 in P. americana were investigated by immunohistochemical staining.
Results
The rPer a 1.0105 (~13 kDa) had 100%, 98% and ≥90% identity to Per a 1.0105, Per a 1.0101, and Cr-PII, respectively. The B-cell epitope of the Per a 1.0105 specific-MAb was located at residues99 QDLLLQLRDKGV110 contained in all 5 Per a 1.01 isoforms and Per a 1.02. The epitope was analogous to the Bla g 1.02 epitope; however, this B-cell epitope was not an IgE inducer. Per a 1.0105 was found in the midgut and intestinal content of American CR but not in the other organs. The amount of the Per a 1 was ~544 ℃g per gram of feces.
Conclusions
The novel Per a 1 B-cell epitope described in this study is a useful target for allergen quantification in samples; however, the specific MAb can be used as an allergen detection reagent. The MAb based-affinity resin can be made for allergen purification, and the so-purified protein can serve as a standard and diagnostic allergen as well as a therapeutic vaccine component. The finding that the Per a 1 is contained in the midgut and feces is useful to increase yield and purity when preparing this allergen.
Periplaneta americana (American cockroach) has a predilection for a tropical climate.1 They inhabit households, offices, and restaurants and are a major source of inhalant indoor allergens.2,3 Exposure to cockroach (CR) allergens cause sensitization to allergic diseases, especially among atopic subjects.4,5 The sensitized subjects develop a Th2-immune response bias with specific Th2-cell proliferation and cytokine production that mediate class switching of specific B cells to IgE-committed B cells. The B cells differentiate into plasma cells that secrete significant amounts of specific IgE. IgE sensitizes mast cells and basophils by fixing their Fc fragments to Fcε receptors on the cell surface.6 Mild to severe allergic morbidity (i.e., allergic rhinitis and asthma) is associated with the re-exposure of sensitized subjects to CR allergens.7 Asthma due to CR allergens is often more severe and prolonged than morbidity caused by other allergens, such as house dust mites and pets.8 Asthmatic attacks may be fatal if not properly treated.
Several P. americana proteins are known to cause human allergies, and Per a 1 is regarded as a major allergen. This protein binds to IgE in sera of 93%-100% of tested CR allergic patients.9,10,11 Per a 1 is an isoallergen that exists in 6 different isoforms, i.e., Per a 1.0101 (accession no. AF072222), Per a 1.0102 (accession no. U78970), Per a 1.0103 (accession no. U69957), Per a 1.0104 (accession no. U69261), Per a 1.0105 (accession no. AY259514.1), and Per a 1.0201 (accession no. U69260). The molecular mass of Per a 1.0105 (with 6x-His tag) was -13.8 which is the minimum sequence of the tandem repeats.10,12 The proteins have 70%-72% amino acid sequence homology with Bla g 1 which is the major German CR (Blattella germanica) allergen.13 Bla g 1 is found in the CR midgut and excreted in feces.14 The attributes for Per a 1 are unknown; therefore, this study used in situ immunohistochemical staining to investigate the anatomical location of Per a 1.0105 (the Per a 1 isoform of P. americana caught in Thailand) and to determine how it reacted with IgE in sera of CR allergic Thai patients.10 A novel Per a 1.0105 specific B-cell epitope was identified by determining phage mimotopic peptides bound to the Per a 1 specific monoclonal antibody (MAb) and the alignment of the phage peptide sequences with the deduced amino acid sequence of recombinant Per a 1.0105 to determine the tentative MAb-bound epitope. Competitive ELISA verified the epitope and its IgE reactivity.
Animal experiments were approved by Siriraj Animal Care and Use Committee (SI-ACUP) no. 003/2554. Blood sample collection from CR allergic patients of the Allergy Clinic of the Department of Otorhinolaryngology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, was approved by the Siriraj Ethical Committee (COA No. Si 231/2011).
Recombinant Per a 1.0105 (rPer a 1.0105) was prepared as described previously with modifications.10 PCR primers used for the amplification of Per a 1.0105 coding sequence were: forward 5'-CGAAGCTTAATTCGGCACGAGGGGAG-3' and reverse primer 5'-ACCTCGAGGGGCAGGCCGAACAAGCT-3'. The restriction sites of HindIII and XhoI (underlined letters) were included in the 2 primer sequences, respectively, to facilitate subsequent gene cloning. The PCR amplicon was cloned into a pGEM®-T-Easy vector, and the recombinant vector was put into JM109 E. coli. The DNA insert was verified by DNA sequencing, and the deduced amino acids were aligned with Per a 1.0105 amino acid sequence (AAP13554.1) by a BLASTp scoring matrix. The verified DNA insert was subcloned into a pET-23b+ protein expression vector. Both the recombinant TA cloning vector that carried the Per a 1.0105 DNA insert and the pET23b+ plasmids were doubly digested with the HindIII and XhoI endonucleases. The cut Per a 1.0105 DNA and the protein expression plasmids were mixed with T4 DNA ligase (New England Biolabs, Ipswich, MA, USA) and maintained at 16℃ for 16 hours before transforming the ligation mixture into E. coli BL21 (DE3) pLysS using a heat-shock method. The transformed preparation was spread onto a Luria-Bertani (LB)-ampicillin (LB-A) agar plate and incubated at 37℃ overnight. A colony of the transformed bacteria from the overnight selective agar plate was inoculated into 5 mL of LB-ampicillin broth and incubated at 37℃ with shaking at 250 rpm overnight. One milliliter of the seed culture was put into 50 mL of fresh LB-ampicillin broth and incubated until the optical density absorbance at 600 nm was 0.6; subsequently, IPTG was added to the final concentration of 1 mM, and the culture was incubated for 3 hours. The bacterial cells were harvested and added to a lysis buffer that contained protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany) and sonicated at 20 kHz, 2 minutes of pulse-on, 3 minutes of pulse-off in ice-bath. Cell debris was removed, and the supernatant was collected. The presence of a protein band (thought to be the recombinant Per a 1.0105 in the lysate) was checked by SDS-PAGE and Coomassie Brilliant Blue G-250 (CBB) staining, Western blot analysis using anti-6×-HisProbe-HRP and substrate, MALDI-TOF/TOF, and database search. Recombinant Per a 1.0105 was purified from the E. coli lysate using pre-equilibrated Ni-NTA resin (ProBond™, Invitrogen, Carlsbad, CA, USA).
Native Per a 1.0105 was prepared from crude P. americana extract as described previously.15 Briefly, CNBr-activated Sepharose™ 4B resin (GE Healthcare, Uppsala, Sweden) was coupled with purified Per a 1.0105 specific-MAb (10 mg of MAb per gram of resin in coupling buffer) according to the instruction manual. After washing with 0.01 M phosphate buffer, pH 7.4 (PB) to remove unbound protein, 7 mL of binding buffer (containing 30 mg of P. americana extract) were mixed with the Affi-Gel on a rotating platform at 4℃ overnight. Unbound proteins were eliminated by washing with PB, then the native Per a 1 was eluted with 0.1 M glycine-HCl buffer, pH 3.0, and neutralized with 1 M Tris-HCl, pH 8.0. Fractions with absorbance at 280 nm were pooled and dialyzed against distilled water at 4℃ overnight. The protein content of the preparation was determined using Bradford reagents (Bio-Rad, Hercules, CA, USA).
Mouse monoclonal antibodies (MAb) specific to Per a 1.0105 were produced as described previously.15 Female BALB/c mouse aged 6 weeks was immunized intraperitoneally with purified rPer a 1.0105 (10 µg in PBS) mixed with alum (Pierce, Rockford, IL, USA) in an antigen to adjuvant ratio of 1:3. Two booster doses were given to the primed mouse at 14-day intervals. One week after the last booster, the mouse was bled, and the serum antibody titer was determined against the homologous protein by indirect ELISA. The mouse was then injected intravenously with 10 µg rPer a 1.0105 in 200 µL PBS. Three days after the intravenous booster, the mouse was bled, and the serum was collected (immune serum to rPer a 1.0105; IS). Splenocytes were fused with P3×-63-Ag8.653 mouse myeloma cells (~10:1) using PEG-4000 as a fusogen. Hybridoma clones secreting MAb reactive only to the homologous antigen and the P. americana whole body extract were established.15
Antigen was separated by 12% SDS-PAGE. It was stained by CBB dye for direct visualization of the protein band(s) or its separated components were electroblotted onto a nitrocellulose membrane (NC; Bio-Rad) for Western blot analysis. For the latter, the empty sites on the blotted NC were blocked with unrelated protein (3% skim milk or BSA in PBS), and the membrane was cut vertically into strips. The NC strips were probed individually with primary antibody, i.e., mouse anti-6×-His antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) to trace the SDS-PAGE-separated rPer a 1.0105 band or hybridoma culture supernatants and to check for the presence/antigenic specificity of the mouse monoclonal antibody to rPer a 1.0105. In both instances, goat anti-mouse IgG-alkaline phosphatase (AP) conjugate and BCIP/NBT chromogenic substrate (KPL, Gaithersburg, MD, USA) were used as reagents to reveal the antigen-antibody reactive band(s).
An indirect ELISA15 determined the mouse serum antibody titer that detected the antibody in the culture fluids of hybrid cells during the selection of hybridoma clones or the reactivity of the MAb (either in culture medium or protein-G purified) to homologous and heterologous antigens. The heterologous antigens used to check the cross-reactivity of the MAb included lysate of E. coli BL21 (DE3), crude extracts of house dust mites (Dermatophagoides pteronyssinus and D. farinae) and crude American CR extract. Each well of the 96-well ELISA plates (Costar, Cambridge, MA, USA) was coated with antigen (0.5 µg in 100 µL carbonate-bicarbonate buffer, pH 9.6) and incubated at 37℃ overnight. Wells were washed with PBS containing 0.05% Tween-20 (PBST), blocked with 1% BSA in PBS and incubated at 37℃ for 1 hour. After another wash, antibody or PBS (blank) was added to the appropriate well and incubated at 37℃ for another 1 hour. Goat anti-mouse total immunoglobulin-horseradish peroxidase (HRP) conjugate (diluted 1:5,000) and ABTS substrate (KPL) were used for color development. The enzymatic reaction was stopped by adding 100 µL of 1% sodium dodecyl sulfate (SDS) to each well. Absorbance at 405 nm of the content of each well was determined against blank using ELISA reader (Multiscan EX Labsystem, Helsinki, Finland).
The isotype of the MAb was determined by ELISA using mouse MAb isotyping reagent (Iso2-1kt, SIGMA, St. Louis, MO, USA) according to the manufacturer's instructions.
Phage peptides (mimotopes) bound to rPer a 1.0105 specific MAb were determined16 using a Ph.D.-12™ (Phage Display Peptide Library Kit; Promega, Madison, WI, USA) which consisted of M13 filamentous phages that displayed a large repertoire of random 12-mer peptides on the surface. A well of a micro-ELISA plate was coated with 1 µg of protein-G purified Per a 1.0105-specific MAb, and the plate was maintained at 4℃ overnight. The unbound protein was washed away by Tris buffered saline (TBS) that contained 0.1% Tween-20 (TBST); each well was blocked with 300 µL of 0.5% BSA in TBS. Peptide display phages (~1.5×1011 plaque forming units; pfu) were added to the MAb-coated well and maintained at 25℃ for 30 minutes. Unbound phages were removed by washing with TBST; the MAb-bound phages were eluted with 0.2 M glycine-HCl, pH 2.2 that contained 1% BSA and propagated in E. coli K12 ER2738. Three rounds of the phage bio-panning were performed. Phages of the third round were transfected with E. coli ER2738, and the preparation was diluted 10-2-10-4 before plating onto top agarose on an LB agar plate that contained IPTG and X-gal. Isolated blue plaques were randomly picked, inoculated individually into 1 mL of LB broth and incubated at 37℃ with shaking at 250 rpm for 4 hours. The bacterial cells were removed by centrifugation and phages in the supernatant were collected by precipitating the culture supernatant with PEG/NaCl. The phage DNA was extracted using phenol/chloroform and absolute ethanol precipitation. DNA coding for the 12-mer peptide of each phage clone was sequenced using M13-96gIII-sequencing primer (New England Biolabs, Hitchin, Herts, UK). Amino acid sequences were deduced. Phage peptides (mimotopes) were then aligned with the rPer a 1.0105 peptide sequence of the present study as well as the rPer a 1.0105 sequences of the NCBI database using ClustalW 2.0.12 multiple sequence alignment to determine the residues of rPer a 1.0105 bound by MAb (epitope).
Per a 1.0105 MAb epitope was verified by competitive ELISA17 with modification. Titers of the mimotopic phages (test) and irrelevant phages (background control) were determined as guided by the instruction manual (New England Biolabs). The test and control phages (100 µL of 5×1010 pfu) were mixed with 5 or 50 µg of rPer a 1.0105-specific MAb. Each preparation was filled into each well of an ELISA plate coated with 1 µg/well of native Per a 1. The antigen-coated well added with diluent of the MAb served as a negative inhibition control or blank. The ELISA procedure was conducted using goat anti-mouse IgG-HRP conjugate (Santa Cruz) and 2,2'-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt substrate (ABTS) (KPL) system. The absorbance at 405 nm of the content in each well was determined against the blank. A lower absorbance at 405 nm of the tests compared to the background and negative inhibition controls implies that the test phages were able to compete for the MAb that validated the phage mimotope and the MAb epitope.
The procedure of competitive ELISA was performed with modification to validate the IgE reactivity of the Per a 1.0105 epitope.17 The amplified rPer a 1.0105-specific MAb mimotopic phages and irrelevant phage (background control) at various amounts (108, 109, 1010, 1011, and 1012 pfu) were mixed individually with pooled CR allergic patient sera (5 patients). Phages mixed with antibody diluents served as a negative inhibition control or blank. The mixtures were added individually into wells of ELISA plate (Corning Inc., Corning, NY, USA) coated with 1 µg/well of native Per a 1 and incubated. After washing, biotinylated goat anti-human IgE (Southern Biotech, Birmingham, AL, USA), streptavidin conjugated with alkaline phosphatase (Southern Biotech) and p-nitrophenyl phosphate, disodium salt (PNPP; Thermo Fisher Scientific, Rockford, IL, USA) were added for color development with washing between steps. The absorbance at 405 nm of the content in each well was measured at against the blank using an ELISA reader (Multiscan EX; Lab systems).
Ten laboratory reared-American CR (5 females and 5 males) were euthanized in CO2 before dissecting their internal organs (salivary glands, gut, muscles, and reproductive tissues). Individual tissues were fixed in 4% paraformaldehyde at 4℃ for 24 hours. The tissues were delipidized, rinsed with water, washed with PBST, dehydrated through graded alcohols, washed with xylene, embedded in molten paraffin and maintained at room temperature overnight. Serial sections (6 µm) of each paraffin block were placed individually on poly-L-lysine coated glass slides. The slides were subjected to immunohistochemical staining by incubating at 55℃ for 30 minutes before removing the paraffin with xylene, rehydrating through reverse-graded alcohol solutions from 100% to 80% and rinsing with distilled water. Endogenous peroxidase was eliminated by incubating the tissues with 3% H2O2 at 37℃ for 15 minutes. After washing, the sections were incubated with 2.5% BSA in PBS at 25℃ for 40 minutes before incubating with Per a 1.0105-specific MAb at 4℃ for 18 hours. After washing away unbound materials with PBST, the sections were incubated with goat anti-mouse total immunoglobulin-HRP conjugate (1:400) at 25℃ for 1 hour. All sections were rinsed in PBST and incubated in 200 µL of 3, 3-diaminobenzidine (DAB)-tetrahydrochloride substrate at 25℃ for 5 minutes. The reaction was stopped by washing with distilled water, mounted with Permount and observed under a light microscope for MAb stained tissues.
Fecal pellets from 10 adult American CRs were collected using a small brush to isolate individual pellets and a forceps to pick them up from the rearing container. The collected feces were pooled, crushed to powder and 0.1 g of the powder was added with 2 mL of PBST. After mixing, the preparation was centrifuged at 5,000×g, 25℃ for 10 minutes; the supernatant was collected to detect the Per a 1.0105 amount by sandwich-ELISA (as described previously).18 Individual wells of an ELISA plate (Corning) were added with 4 µg of purified Per a 1.0105 specific-MAb in 100 µL carbonate-bicarbonate buffer, pH 9.6, and the plate was maintained overnight at 37℃ in a humid chamber. The wells were washed with PBST and blocked with 1% BSA in PBST; subsequently, 100 µL of the CR fecal extract (diluted 1:2,000, 1:4,000 and 1:8,000 in PBST) were added appropriately to the MAb coated wells and incubated at 37℃ for 30 minutes. After washing, 100 µL of 1:1,000 rabbit polyclonal antibodies to American CR extract were added to each well and the plate was re-incubated at 37℃ for 1 hour. Goat anti-rabbit total immunoglobulin-HRP conjugate (100 µL of 1:1,000) and ABTS substrate (KPL) were used for color development. The enzymatic reaction was stopped by adding 100 µL of 1% SDS. The absorbance at 405 nm of the content in each well was measured against blank (PBS was added to the MAb-coated wells instead of the fecal sample). The amount of the Per a 1.0105 in the fecal sample was calculated from a standard curve constructed from the absorbance at 405-nm readouts of the known concentrations of recombinant Per a 1.0105 subjected similarly to the sandwich ELISA and multiplied by the dilution factor.
Multiple alignments (BLASTn) of the gene sequence coding for the Per a 1.0105 of this study with the Per a 1.0105 (AY259514.1), Per a 1.0101 (AP0722222.1), and Cr-PII (U78970.1, U59957.1, U69261.1 and GU01383.1) showed 100%, 98%, and ≥90% identity, respectively. Per a 1.0105 shared a 77% nucleotide sequence identity with the Bla g 1.02 of Blattella germanica. Fig. 1A and B shows recombinant Per a 1.0105 (-13 kDa) purified from lysate of a selected transformed BL21 (DE3) E. coli clone in SDS-PAGE gel stained by the CBB dye and Western blot patterns of the protein probed with mouse monoclonal anti-6×His antibody, goat anti-mouse IgG-AP conjugate, and AP-substrate, respectively. The protein band was identified by MALDI-TOF/TOF as Per a 1.0105 (17 peptides of the produced rPer a 1.0105 matched the peptides of Per a 1 [accession no. gi30144660] with a sequence coverage of 68% and a total score of 769).
Monoclonal antibodies from 2 hybridoma clones (ACR1 and ACR2) reacted with rPer a 1.0105 and a whole body extract of P. americana; however, they did not react to other heterologous antigens, such as E. coli BL21 (DE3) lysate and crude extracts of D. pteronyssinus and D. farinae as determined by indirect ELISA. The isotype of immunoglobulin of both clones was IgG1. Fig. 2 shows the Western blot pattern of the SDS-PAGE separated lysate of E. coli BL21 (DE3) carrying the Per a 1.0105-pET-23b+ probed with the culture supernatants of hybridoma clones ACR1 and ACR2 (containing MAbACR1 and MAbACR2, respectively). The titer of the produced MAb from clone ACR1 was higher than ACR2; therefore, the MAb produced by the ACR1 clone was used in subsequent experiments.
Only 6 from 20 randomly picked phage clones from panning with the purified MAbACR1 revealed peptides matched with the amino acid sequence of Per a 1.0105 (accession no. AY259514.1). The pairwise alignments of a selected phage peptide, REVLQKNFAKGV, with Per a 1.0105 amino acid sequence showed that the phage, mimotope matched residues between 99QDLLQNLRDKGV110 of the CR allergen (Fig. 3). Complete matches were at the amino acids102 LQ----KGV;108 however, the amino acids at positions 99-101 and 104 were conserved amino acids. Therefore, the region of Per a 1.0105 that matched the phage mimotope derived from panning with MAbACR1 should be a presumptive Per a 1.0105 epitope of the MAbACR1.
Fig. 4 shows the results of the competitive ELISA that determined the ability of phage mimotopes to inhibit the MAb binding to the native Per a 1. The binding of 5 and 50 µg of the rPer a 1.0105 specific-MAb to the native Per a 1 was significantly inhibited (42% and 36%, respectively) by phage mimotopes (phages carrying peptides that bound to the MAb); subsequently, this implies that the phage carried amino acid residue analogous to the native Per a 1 and validated the mimotope search results.
The result of IgE ELISA inhibition assay revealed that the phage mimotope at amounts of 108, 109, 1010, 1011, and 1012 pfu performed similarly to the irrelevant phage; however, both could not inhibit the binding of the IgE in patient-pooled sera to the native Per a 1 and indicated that the MAb-specific Per a 1 epitope was not an IgE-inducing epitope.
Tissue sections of the American CR positive for the Per a 1.0105 specific-MAb based-immunohistochemical staining showed a brownish-yellow color under a light microscope. An examination of all processed sections revealed that only the mucosa of the midgut and gut content were positive for staining (Fig. 5); no Per a 1 immunoreactivity was observed in the other internal organs (salivary gland, gut, hemolymph, testes/ovary and muscle sections). There was no noticeable difference between male and female CR sections in staining patterns and intensity. Per a 1 immunoreactivity was not detected in any control sections incubated with normal mouse serum instead of MAb.
A high asthma prevalence (40%-60%) of patients with IgE to cockroach allergens is reported in several Thai cities. An epidemiological survey of 14 provinces in Thailand showed that American CR (P. americana) is the predominant household domiciliary CR.19 This is related to the incidence of CR allergy in Thailand.20 Per a 1.0105 (a variant of Per a 1 from P. americana) is identified as one of the major allergens as the protein reacted with IgE in sera of all Thai CR allergic subjects.10 Per a 1 comprises 2 isoallergens, i.e., Per a 1.01 and Per a 1.02. Five variants of the Per a 1.01, i.e., Per a 1.0101, Per a 1.0102, Per a 1.0103, Per a 1.0104, and Per a 1.0105, have been recognized.11 In this study, mouse MAb specific to Per a 1.0105 was produced and used to determine Per a 1.0105-specific B-cell epitope as well as locate the source of the Per a 1 in American CR.
Recombinant Per a 1.0105 (rPer a 1.0105) is produced from E. coli carrying recombinant Per a 1.0105-pET23b+ plasmids, which had a strong promoter and provided a high yield of recombinant protein compared to Per a 1.0105-pPIC9 plasmids.10 The rPer a 1.0105 is found in the soluble part; therefore, the molecule should acquire a native configuration. The soluble protein in monomeric form (13 kDa)10 is readily purified from the E. coli lysate by an affinity resin and is confirmed to be Per a 1.0105 by SDS-PAGE, immunoblotting and mass spectrometry.
Mouse MAb specific to the rPer a 1.0105 is successfully produced using conventional hybridoma technology. The so-produced MAb (IgG1 isotype) can serve as a reagent for the detection and quantification of the American CR allergen in an allergy intervention measure. In this study, specific MAb was used for the identification of Per a 1-specific B-cell epitope and to reveal in situ locations of the allergen.
B-cell epitopes (that included specific IgE-binding regions of the Per a 1) have been studied previously using Per a 1.0104 deletion mutants and synthetic overlapping peptide inhibition of patient serum IgE binding.21 It has been found that IgE in atopic human sera bound to heterogeneous epitopes span on the Per a 1.0104 molecule. The amino acids 78LIRALFGL85 and 267IRSWFGLP274 located in the internal repeats of the Per a 1.0104 variant show 80% and 100% positive IgE binding. In this study, the B-cell epitope of the Per a 1.0105 bound to the MAb was readily identified using a 12-mer random peptide phage display library to select phages that displayed peptides that mimic the original characteristic of the native epitope of the Per a 1.0105-specific MAb (epitope).22 Validity of this technique has been demonstrated previously in several studies.17,23,24,25 The phage mimotopic peptide is REVLQKNFAKGV which is matched with the amino acids 99QDLLQNLRDKGV110 of the Per a 1.0105. The REVLQK----KGV of the phage peptide and the 99QDLLQ----KGV110 of the Per a 1.0105 sequence are conserved/identical amino acids; however, the other 4 amino acids (KNFA versus NLRD) were not matched. This explains the incomplete inhibition of MAb binding to the native Per a 1.0105 by the phage mimotope in the competitive ELISA; in addition, the amounts of the MAb in the competitive assay might be excessive. This B-cell epitope did not bind to IgE in the sera of patients and was consistent with the previous finding that human atopic sera did not recognize amino acids 1-77, 86-205, and 200-266 of the Per a 1.0104 allergen.21 However, multiple alignments indicated that this non-IgE binding site is shared by 4 other variants of Per a 1.01 that include Per a 1.0101, Per a 1.0102, 1.010, and Per a 1.0104 as well as Per a 1.0201. The epitope sequence is analogous to amino acids 465ILQRLKDKG474 of the Bla g 1 (accession no. EF202179).26 Western blot analysis revealed that the MAb bound to recombinant Bla g 1 (Supplementary data 2); subsequently, the Per a 1.0105 specific-MAb produced in this study has potential use as a broad capture reagent for the detection and quantification of American CR allergen in samples as well as the B. germanica major allergen (Bla g 1). MAb can be used to prepare affinity resin for specific capturing of native Per a 1 and Bla g 1 from respective crude CR extracts. Native allergens in pure form can be used as specific allergens to monitor CR allergic patient status and standard reagents in allergen quantification test kits. They may also be used as refined and more effective therapeutic CR allergy vaccine components (especially to reduce airway inflammation) versus the crude CR extracts demonstrated recently.27
MAb to rPer a 1.0105 was utilized in immunochemical staining to locate the allergen in various tissues and organs of P. americana using polyclonal antibodies of the rPer a 1.0105 as a positive control. A previous study showed that the deduced amino acid sequence of the rPer a 1.0105 had hydrophilic and hydrophobic regions in the molecule, which suggest it is a transmembrane protein. The protein is analogous to a putative protein that involves digestion in the midgut (ANG12 precursor) Aedes aegypti.10,13 Per a 1 and Bla g 1 showed an amino acid sequence identity of 70%-72%; in addition, both proteins are antigenically cross-reactive.13 Bla g 1 (found in B. germanica proventriculus and hindgut) has been shown to be induced after food intake.13 In this study, a strong positive signal for the Per a 1.0105 was also found in the epithelium of the insect midgut that signified that the allergen was generated from the midgut of American CRs and excreted in relatively high amounts in feces (-544 µg per gram of the CR feces). Both Per a 1 and Bla g 1 may have an important function in relation to the peritrophic membrane which is a layer of chitin and proteins that encircle the insect gut, i.e., prevent the gut epithelium from damage caused by abrasive food bolus and pathogens, such as viruses, bacteria, protozoa, and helminthes.28 The proteins may play a part or sole protective role in the assimilation and elimination of toxic ions.29 In mosquitoes, the peritrophic membrane is proven to protect the gut from toxic heme derivatives after taking the blood meal.30 The speculative roles of Bla g 1 and Per a 1 await experimental validation.
This study provides initial information on the source of Per a 1 allergen of P. americana. A novel immunodominant B-cell epitope shared by all Per a 1.01 variants, Per a 1.02 as well as Bla g 1 of B. germanica, was revealed by means of phage mimotope searching and multiple alignments. The epitope is a useful target of the allergen quantification test kit, which is a necessary tool for CR allergy intervention. The so-produced Per a 1.0105-specific MAb may be used as an affinity reagent to capture the native protein from the crude CR extract for further use as a refined allergen to monitor the allergic status of a patient or as a therapeutic component of a CR allergy vaccine.
Figures and Tables
Fig. 1
SDS-PAGE separated-rPer a 1.0105 stained by Coomassie Brillian blue G-250 dye (A) and probed with mouse monoclonal anti-6x-histidine antibody (B). Lanes M of both blocks, Protein molecular weight standards. Numbers at the left of both blocks are molecular masses of proteins in kDa.

Fig. 2
Western blot results for determining antigenic specificity of the produced MAb. Lanes 1 and 2, SDS-PAGE separated-rPer a 1.0105 blotted strips probed with culture supernatants of hybridoma clones ACR1 and ACR2, respectively. Lanes 3 and 4, SDS-PAGE separated E. coli BL21 (DE3) lysate blotted strips probed with culture supernatants of hybridoma clones ACR1 and ACR2, respectively (controls). Lane M, Protein molecular weight standards. Numbers at the left are protein molecular masses in kDa.
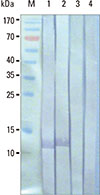
Fig. 3
Pairwise alignments of the phage mimotope peptide (MAbPer a 1.0105-1) with the Per a 1.0105 amino acid sequence for the identification of the Per a 1.0105 residues bound by the ACR1 MAb.
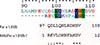
Fig. 4
Results of competitive ELISA for verification of the MAb epitope. The tested phages carrying the MAb mimotope (peptide that bound to the MAb) could inhibit the binding of 5 and 50 µg of MAb to native Per a 1.0105 compared with the irrelevant phages and diluent controls. # and *, different significantly from the inhibition by the irrelevant phages at 5 and 50 µg of MAb, respectively.

ACKNOWLEDGMENTS
The work was supported by the National Research University (NRU) project of the Office of Commission on Higher Education, Ministry of Education, Thailand through the Center of Biopharmaceutical Development and Innovative Therapy, Mahidol University and DPG5380001 of the Thailand Research Fund (TRF). Anchalee Tungtrongchitr and Nitat Sookrung are supported by a "Chalermphrakiat" Grant, Faculty of Medicine Siriraj Hospital, Mahidol University. Nitat Sookrung and Nitaya Indrawattana are new research scholars of the TRF. The authors thank all CR allergic patients for their participation in this study.
References
1. Gore JC, Schal C. Cockroach allergen biology and mitigation in the indoor environment. Annu Rev Entomol. 2007; 52:439–463.
2. Ahluwalia SK, Matsui EC. The indoor environment and its effects on childhood asthma. Curr Opin Allergy Clin Immunol. 2011; 11:137–143.
3. Cohn RD, Arbes SJ Jr, Jaramillo R, Reid LH, Zeldin DC. National prevalence and exposure risk for cockroach allergen in U.S. households. Environ Health Perspect. 2006; 114:522–526.
4. Huss K, Adkinson NF Jr, Eggleston PA, Dawson C, Van Natta ML, Hamilton RG. House dust mite and cockroach exposure are strong risk factors for positive allergy skin test responses in the Childhood Asthma Management Program. J Allergy Clin Immunol. 2001; 107:48–54.
5. Matsui EC, Wood RA, Rand C, Kanchanaraksa S, Swartz L, Curtin-Brosnan J, Eggleston PA. Cockroach allergen exposure and sensitization in suburban middle-class children with asthma. J Allergy Clin Immunol. 2003; 112:87–92.
6. Chew GL, Perzanowski MS, Canfield SM, Goldstein IF, Mellins RB, Hoepner LA, Ashby-Thompson M, Jacobson JS. Cockroach allergen levels and associations with cockroach-specific IgE. J Allergy Clin Immunol. 2008; 121:240–245.
7. Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, Mitchell H, McNiff-Mortimer K, Lynn H, Ownby D, Malveaux F. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. N Engl J Med. 1997; 336:1356–1363.
8. Chapman MD, Vailes LD, Hayden ML, Platts-Mills TA, Arruda LK. Cockroach allergens and their role in asthma. In : Kay AB, editor. Allergy and allergic diseases. Oxford: Blackwell Science;1997. p. 942–951.
9. Schou C, Lind P, Fernandez-Caldas E, Lockey RF, Løwenstein H. Identification and purification of an important cross-reactive allergen from American (Periplaneta americana) and German (Blattella germanica) cockroach. J Allergy Clin Immunol. 1990; 86:935–946.
10. Diraphat P, Sookrung N, Chaicumpa W, Pumhirun P, Vichyanond P, Tapchaisri P, Kalambaheti T, Mahakunkijchareon Y, Sakolvaree Y, Bunnag C. Recombinant American cockroach component, Per a 1, reactive to IgE of allergic Thai patients. Asian Pac J Allergy Immunol. 2003; 21:11–20.
11. Sookrung N, Chaicumpa W. A revisit to cockroach allergens. Asian Pac J Allergy Immunol. 2010; 28:95–106.
12. Wu CH, Luo SF, Wong DW. Analysis of cross-reactive allergens from American and German cockroaches by human IgE. Allergy. 1997; 52:411–416.
13. Pomés A, Wünschmann S, Hindley J, Vailes LD, Chapman MD. Cockroach allergens: function, structure and allergenicity. Protein Pept Lett. 2007; 14:960–969.
14. Gore JC, Schal C. Gene expression and tissue distribution of the major human allergen Bla g 1 in the German cockroach, Blattella germanica L. (Dictyoptera: Blattellidae). J Med Entomol. 2004; 41:953–960.
15. Sookrung N, Diraphat P, Chaicumpa W, Tongtawe P, Sakolvaree Y, Tapchaisri P, Mahakittikun V, Tungtrongchitr A, Vichyanond P, Bunnag C. Cockroach allergen detection and cockroach allergens of allergic Thai patients. Asian Pac J Allergy Immunol. 2003; 21:1–9.
16. Kulkeaw K, Sakolvaree Y, Srimanote P, Tongtawe P, Maneewatch S, Sookrung N, Tungtrongchitr A, Tapchaisri P, Kurazono H, Chaicumpa W. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, neurotoxin. J Proteomics. 2009; 72:270–282.
17. Thueng-in K, Thanongsaksrikul J, Srimanote P, Bangphoomi K, Poungpair O, Maneewatch S, Choowongkomon K, Chaicumpa W. Cell penetrable humanized-VH/V(H)H that inhibit RNA dependent RNA polymerase (NS5B) of HCV. PLoS One. 2012; 7:e49254.
18. Tungtrongchitr A, Sookrung N, Chaicumpa W, Indrawattana N, Meechan T, Thavara U, Visitsunthorn N, Bunnag C. Comparison of allergenic components and biopotency in whole body extracts of wild and laboratory reared American cockroaches, Periplaneta americana. Asian Pac J Allergy Immunol. 2012; 30:231–238.
19. Tawatsin A, Thavara U, Chompoosri J, Kong-ngamsuk W, Chansang C, Paosriwong S. Cockroach surveys in 14 provinces of Thailand. J Vector Ecol. 2001; 26:232–238.
20. Tungtrongchitr A, Sookrung N, Munkong N, Mahakittikun V, Chinabut P, Chaicumpa W, Bunnag C, Vichyanond P. The levels of cockroach allergen in relation to cockroach species and allergic diseases in Thai patients. Asian Pac J Allergy Immunol. 2004; 22:115–121.
21. Wu CH, Lee MF, Yang JS, Tseng CY. IgE-binding epitopes of the American cockroach Per a 1 allergen. Mol Immunol. 2002; 39:459–464.
22. Sanderson SD, Cheruku SR, Padmanilayam MP, Vennerstrom JL, Thiele GM, Palmatier MI, Bevins RA. Immunization to nicotine with a peptide-based vaccine composed of a conformationally biased agonist of C5a as a molecular adjuvant. Int Immunopharmacol. 2003; 3:137–146.
23. Sookrung N, Chaicumpa W, Tungtrongchitr A, Vichyanond P, Bunnag C, Ramasoota P, Tongtawe P, Sakolvaree Y, Tapchaisri P. Periplaneta americana arginine kinase as a major cockroach allergen among Thai patients with major cockroach allergies. Environ Health Perspect. 2006; 114:875–880.
24. Thanongsaksrikul J, Srimanote P, Maneewatch S, Choowongkomon K, Tapchaisri P, Makino S, Kurazono H, Chaicumpa W. A V H H that neutralizes the zinc metalloproteinase activity of botulinum neurotoxin type A. J Biol Chem. 2010; 285:9657–9666.
25. Chavanayarn C, Thanongsaksrikul J, Thueng-In K, Bangphoomi K, Sookrung N, Chaicumpa W. Humanized-single domain antibodies (VH/VHH) that bound specifically to Naja kaouthia phospholipase A2 and neutralized the enzymatic activity. Toxins (Basel). 2012; 4:554–567.
26. Yi MH, Jeong KY, Kim CR, Yong TS. IgE-binding reactivity of peptide fragments of Bla g 1.02, a major German cockroach allergen. Asian Pac J Allergy Immunol. 2009; 27:121–129.
27. Meechan P, Tungtrongchitr A, Chaisri U, Maklon K, Indrawattana N, Chaicumpa W, Sookrung N. Intranasal, liposome-adjuvanted cockroach allergy vaccines made of refined major allergen and whole-body extract of Periplaneta americana. Int Arch Allergy Immunol. 2013; 161:351–362.
28. Lehane MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997; 42:525–550.
29. Kato N, Dasgupta R, Smartt CT, Christensen BM. Glucosamine: fructose-6-phosphate aminotransferase: gene characterization, chitin biosynthesis and peritrophic matrix formation in Aedes aegypti. Insect Mol Biol. 2002; 11:207–216.
30. Pascoa V, Oliveira PL, Dansa-Petretski M, Silva JR, Alvarenga PH, Jacobs-Lorena M, Lemos FJ. Aedes aegypti peritrophic matrix and its interaction with heme during blood digestion. Insect Biochem Mol Biol. 2002; 32:517–523.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download