Aspirin ingestion can induce a wide range of clinically recognized allergic reactions, including acetyl salicylic acid (ASA)-exacerbated respiratory disease (AERD), ASA-intolerant urticaria (AIU), chronic rhinitis, and anaphylaxis. Patients with AIU have high rates of atopy and increased total IgE levels,
1 but the pathogenic mechanism of AIU remains unclear. ASA is metabolized by UDP-glucuronosyltransferase 1A6 (UGT1A6), cytochrome P4502C9 (CYP2C9), and N-acetyl transferase 2 (NAT2). Inter-individual differences in the activities of these enzymes may be the underlying cause of adverse ASA-related symptoms such as urticaria.
The two polymorphic enzymes CYP2C9 and UGT1A6 are involved in hydroxylation and glucuronidation of ASA, respectively.
2,
3 Two known variant alleles of
UGT1A6 result in amino acid changes at positions 181 and 184, and a 30-50% reduction in enzyme activity compared with wild type activity.
4 Similarly, two variant
CYP2C9 alleles,
CYP2C9*2 (R144C) and
CYP2C9*3 (I359L), produce slow metabolizing enzymes, with 5-30% of wild-type enzyme activity.
5 Variations in the enzymes that metabolize ASA may play a role in ASA-related diseases such as urticaria.
Leukotriene overproduction is a possible risk factor for ASA hypersensitivity, including AIU.
6 Cysteinyl leukotrienes are inactivated by acetyl coenzyme A-dependent N-acetyltransferase (NAT). Thus, functional alterations in the
NAT2 gene may contribute to the risk for ASA-intolerant symptoms such as asthma.
7 Genetically-determined rapid and slow acetylators produce variation in the elimination rates of xenobiotics, as well as in the levels of NAT2 in the liver and other tissues. Single nucleotide polymorphisms (SNPs) of the
NAT2 gene are markers of atopic asthma, high serum IgE levels, and high blood eosinophil counts.
8
Based on the involvement of these enzymes in the metabolism of ASA, we investigated the relationship between polymorphisms in
UGT1A6,
CYP2C9, and
NAT2, and the occurrence of ASA-intolerant urticaria in a Korean population. In total, 148 AIU patients (AIU group) and 260 normal healthy control subjects (NC) were enrolled at the Ajou University Hospital in Suwon, Korea. AIU was defined by a positive result on an ASA oral provocation test, which was performed using 500 mg of ASA (Rhonal; KunWha Pharmaceutical Co., Seoul, Korea).
5 A control group (NC) with no personal or family history of allergic diseases or past history of ASA hypersensitivity was recruited from the general population. Informed consent was obtained from all subjects prior to enrollment, and the study was approved by the Ajou University Hospital Institutional Review Board. Skin prick testing was performed with 55 common aero-allergens (Bencard Co., West Sussex, UK). Atopy was defined as one or more positive reactions to common inhalant allergens. Total IgE concentration was measured using a UniCAP system (Pharmacia Diagnostics, Uppsala, Sweden), according to the manufacturer's instructions. SNPs in the promoter and/or exons of the
CYP2C9,
UGT1A6, and
NAT2 genes of 40 healthy volunteers were sequenced using an ABI Prism 3100 DNA analyzer (Applied Biosystems, Foster City, CA, USA). We identified two SNPs in
CYP2C9, one in the promoter (
CYP2C9 -1188T>C) and one in an exon (
CYP2C9*3A1075C). For
UGT1A6, we selected two exonic SNPs (
UGT1A6 T181A A>G and
UGT1A6 R184S A>C). We confirmed five SNPs for
NAT2 (
NAT2 9796A>T,
NAT2 197G>A,
NAT2 286G>A,
NAT2 9601A>G, and
NAT2 9306A>G). These SNPs were genotyped using primer extension and a SNAPshot ddNTP primer extension kit (Applied Biosystems).
Differences in mean values of phenotypic characteristics among the patients were compared using a Student's t-test for continuous variables or the χ2 test for categorical variables. Differences in genotype frequency between the two groups were examined using the χ2 test. A logistic regression recessive analysis model was used after adjustment for age and gender covariates, followed by a multiple comparison test. All statistical analyses were performed using SPSS version 12.0 (SPSS Inc., Chicago, IL, USA). In all analyses, P<0.05 was considered statistically significant.
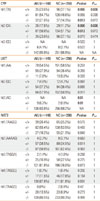
The mean age was significantly higher in the AIU group than in the NC group (35.1±11.5 vs. 32.5±12.6 years, respectively; P=0.041). The AIU patients exhibited a significantly higher rate of atopy than the NC group (69.6 vs. 13.2%, respectively; P<0.001) and higher serum total IgE (log transformed; 5.1±1.2 vs. 3.6±1.3 IU/mL respectively; P<0.001).
The genotype frequencies for the two polymorphisms in each of
CYP2C9 and
UGT1A6 and the five polymorphisms in
NAT2 were analyzed and compared between AIU patients and NC subjects. The two polymorphisms in
CYP2C9 were in linkage disequilibrium (/D'/=1, r
2=0.067). The frequency of the minor C allele of
CYP2C9 -1188T>C was significantly higher in the AIU group than in the NC group (
P=0.005), and remained significant after a multiple comparison (
Pcorr=0.03). The frequency of the variant genotype CC of
CYP2C9 -1188T>C was also higher in the AIU patients compared with the controls in both the co-dominant (
P=0.007) and recessive models (
P=0.012). In addition, the relationship between the
CYP2C9 -1188T>C polymorphism and AIU remained significant after a multiple comparison test (
Pcorr=0.042 [co-dominant model] for AIU vs. NC;
Table 1). Using Power Analysis and Sample Size (PASS, 2008), the power of this genetic association study was calculated to be 74% for detecting an effect size (W) of 0.1430 using 2 degrees of freedom with the χ
2 test, at a significance level (alpha) of 0.05. The frequency of haplotype 2 [CA] was significantly higher in the AIU group compared with the NC group in both the co-dominant (
P=0.006;
Pcorr=0.036) and dominant models (
P=0.012) (
Table 2).
There was no significant difference in allele, genotype, or haplotype frequencies of the UGT1A6 and NAT2 genetic polymorphisms between the AIU and NC groups. Clinical parameters, including atopic status, serum total IgE levels, and autoantibody levels, did not differ between any of the genotypes for any of the polymorphisms.
We analyzed the genotype frequencies for two SNPs in both CYP2C9 and UGT1A6, and five SNPs in NAT2, and compared them between AIU patients and normal control subjects in a Korean population. This is the first study to investigate the SNPs of CYP2C9, UGT1A6, and NAT2 in AIU patients. Among the nine polymorphisms in the three metabolic enzyme genes analyzed, CYP2C9 -1188T>C showed a significant association with the AIU phenotype. The frequencies of the minor C allele and the CC genotype of CYP2C9 -1188T>C were significantly higher in the AIU group compared with the NC group. There was no significant difference in allele, genotype, or haplotype frequencies of the UGT1A6 and NAT2 polymorphisms between the AIU and NC groups.
CYP2C9 is involved in the metabolism of many therapeutic agents, including non-steroidal anti-inflammatory drugs, oral anticoagulants, and angiotensin receptor antagonists. The metabolism of non-steroidal anti-inflammatory drugs involves oxidation by CYP enzymes and conjugation, particularly glucuronidation, by phase II enzymes. Aspirin is deacetylated to salicylic acid and is further metabolized by glucuronidation, hydroxylation, and glycine conjugation, with CYP2C9 playing a major role in the metabolic process.
5 Genetic polymorphisms in
CYP2C9 have been associated with cutaneous adverse reactions induced by diphenylhydantoin.
9 The effects of inter-individual differences in
UGT1A6 and
CYP2C9 genotypes on ASA metabolism have been described in colon adenoma.
5 Recently,
CYP2C19 and
CYP2C9 genetic polymorphisms were shown to be significantly associated with an increased risk for the development of antituberculosis drug-induced maculopapular eruption in the Korean population.
10 Collectively, these findings suggest that the C allele may be involved in the pathogenesis of AIU; however, further studies of CYP2 enzyme activity are needed.
The
NAT2 gene variants and its subphenotypes have been linked to the development of asthma. Additionally, the NAT2 slow acetylator phenotype may be a determinant in a patient's susceptibility to asthma.
11 Another study indicated that SNPs in the
NAT2 gene are closely associated with asthma-related traits such as high serum IgE and blood eosinophil counts, and aspirin hypersensitivity in asthmatic subjects.
7,
8 Significant associations between
UGT1A6 T181A gene polymorphisms and asthma susceptibility have also been reported.
12 However, the results of this study did not show a relationship between
NAT2 or
UGTA6 polymorphisms and AIU in a Korean population.
In summary, we identified a statistically significant association between the C allele of CYP2C9 -1188T>C and AIU, suggesting that this allele may modulate the risk for, and contribute to, the development of the AIU phenotype.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download