Abstract
Major classes of medication in asthma management include bronchodilating β2-agonists, anti-inflammatory inhaled corticosteroids, leukotriene modifiers and theophyllines. However, all asthmatics do not respond to the same extent to a given medication. Available data suggest that a substantial range of individual variability, as much as 70%, may be due to genetic characteristics of each patient. Pharmacogenomics offers the potential to optimize medications for individual asthmatics by using genetic information to improve efficacy or avoid adverse effects. The best-studied case of the potential contribution of pharmacogenomics to treatment response in asthma comes from studies on human β2 adrenergic receptors. In addition, genetic variation in β2-adrenergic receptor (Arg16Gly) may predict response to anticholinergics for the treatment of asthma. In case of inhaled corticosteroids, a recent investigation using a traditional SNP-based approach identified a gene for corticotropin releasing hormone receptor 1 as a potential marker of response. Another major pathway that has been investigated is the pathway underlying response to cysteinyl leukotriene receptor antagonist. It is likely that in the near future, pharmacogenomic approaches based on individual genetic information will be introduced into an asthma treatment guideline and this guideline will allow us to identify those who have the best chance to respond to a specific medication.
Asthma is a chronic inflammatory disorder of the airways characterized by bronchial hyperresponsiveness, reversible airflow limitation, and respiratory symptoms. Asthma is a common disease worldwide, affecting more than 300 million individuals in the developed world.1 The prevalence of asthma varies widely among countries, ranging from 2.1% to 4.4% in developing countries such as Albania, China, and Indonesia to 29.1% to 32.2% in Australia, New Zealand, and the United Kingdom.2 In Korea, the prevalence of asthma in elementary school children increased remarkably from 5.7% in 1981 to 17.0% in 1995 and decreased slightly thereafter (13.0% in 2000 and 10.5% in 2006).3,4 In 2001, the prevalence of asthma tended to be less in adults than in children (3.6%). However, among the elderly (aged 65 years or older), asthma prevalence was high at 12.7%.5 Additionally, the high prevalence of asthma comes at a high cost. According to a study that used the Korea National Health Insurance claims database, in 2004, the estimated total annual cost of asthma was more than $4 billion in Korea [unpublished data].
For this reason, many studies have been conducted to determine more effective treatments for asthma, and major advances in asthma medications have been achieved. However, significant heterogeneity in the efficacy and adverse reactions of asthma therapeutics has been observed, and efforts have been made to clarify the potential genetic sources of this individual variability. The purpose of this review was to present an overview of pharmacogenomics for current asthma treatment.
The goal of asthma treatment is to achieve and maintain clinical control. Asthma treatment can be divided into four main categories: the avoidance of risk factors that cause asthma exacerbations such as allergens, viral infections, pollutants, environmental control to reduce allergens, immunotherapy, and pharmacotherapy. Pharmacotherapy plays a main role in controlling and preventing asthma. The current treatment guidelines recommend a stepwise approach to pharmacologic therapy based on asthma control (Fig. 1).1
Medications to treat asthma can be classified as controllers or relievers. Controllers are medications taken daily on a long-term basis to keep asthma under clinical control chiefly through their anti-inflammatory effects. Controlling medications include inhaled corticosteroids (ICSs), long-acting β2-agonists (LABAs), leukotriene modifiers, methylxanthines and omalizumab. Relievers are medications used on an as-needed basis, which act quickly to reverse bronchoconstriction and relieve its symptoms. Short-acting β2-agonists (SABAs), systemic glucocorticosteroids and short-acting anticholinergics are included as relievers.
The achievement of treatment goals is less than optimal in many patients. Even in patients with an apparently identical clinical phenotype, the response to drug treatment may vary remarkably. Commonly, some patients respond to a given treatment, while others fail to respond. Variability in the individual asthma treatment response may be due to many factors, including the severity and type of disease, treatment compliance, concurrent illness, other medications taken (drug-drug interactions), environmental exposures, and age. However, up to an estimated 70% of the variability in therapeutic responses to pharmacotherapy might be due to genetic variability. Results of a randomized, stratified, double-blind, parallel-group study of 3,421 patients with uncontrolled asthma showed that even treatment that is fully consistent with current guidelines fails to control asthma in up to 25% of patients.6 A study on treatment response to glucocorticosteroids, β2 adrenergic agonists and an experimental leukotriene inhibitor found that 60-80% of the variance in drug response may be due to individual genetic differences.7 This value corresponds to the maximum limit of genetic variance, and indicates that a clinically relevant part of the response to the main classes of asthma drugs may be due to genetic determinants. The genetic variations include a polymorphism in a gene or a random DNA position (single nucleotide polymorphism, SNP) or variations in a series of associated alleles (haplotypes, linkage disequilibrium) and may be related to drug metabolism (pharmacokinetics) or to the drug target (pharmacodynamics). These results also suggest that current treatment guidelines for asthma therapy should be reviewed considering the latest information about genetic determinants of responses to commonly used asthma therapies.
Pharmacogenomics is the study of how inheritance influences a drug response. A greater understanding of the genetic determinants of the drug response may lead to improved patient treatment. By predicting patients at risk for severe toxicity, or those likely to benefit from a particular treatment based on a patient's genetic profile, pharmacogenomics can realize the ultimate goal of individualized asthma therapy. Furthermore, pharmacogenomics offers a new tool for discovering new targets for drug development. To date, asthma pharmacogenomic investigations have focused on β2-agonists, leukotriene antagonists, and glucocorticosteroids. In addition to the pharmacogenomic evidence for these three medications, our pharmacogenomic data on anticholinergics will be presented.
β2-Agonists represent the most important bronchodilator drugs used for treating acute asthma. Because of the many reports on individual differences in response to β2-agonists and tachyphylaxis, investigators have focused on the β2-adrenergic receptor (β2AR) gene. Responses to this drug are currently the most investigated pharmacogenomic approaches in asthma.
The β2AR is a G protein-coupled receptor with seven transmembrane segments and 413 amino acids. Activation of β2AR with β2-agonist binding uncouples stimulatory G proteins, followed by activation of adenyl cyclase, which converts ATP to cAMP and activates protein kinase A.8 Increased cAMP levels and phosphorylation of proteins by protein kinase A indirectly cause relaxation of bronchial smooth muscle and mast cell stabilization.
β2-Agonist pharmacogenomic studies have focused on the effect of ADRB2 variants on the response or adverse outcomes of β2-agonists. ADRB2, encoding β2AR, contains 2,011 nucleotide base pairs on the long arm of chromosome 5 (5q31-32). Among 10 identified functional SNPs, three coding polymorphisms located at positions 16 (rs1042713), 27 (rs1042714) and 164 (rs 1800888) have been studied. These SNPs result in arginine substitution for glycine (Arg16→Gly), glutamine substitution for glutamic acid (Gln27→Glu) and threonine substitution for isoleucine (Thr164→Ile). The Arg16→Gly and Gln27→Glu polymorphisms are frequent in Asians, however, Thr164→Ile polymorphism is only and rarely observed in European Americans.9
Our clinical study of 195 children showed that the bronchodilating response was significantly higher in subjects homozygous for Arg16 than in those homozygous for Gly16 (62.8% versus 46.7%, respectively; P=0.04). Moreover, patients with the homozygous Arg16Gln27 haplotype pairs and those heterozygous for Arg16Gln27/Gly16Glu27 showed the highest bronchodilating responses (62.8% and 61.6%, respectively), and the patients with the homozygous Gly16Gln27 haplotype pairs showed the lowest response (43.2%).11 Similarly, results from one study showed that albuterol-evoked forced expiratory volume in 1 s (FEV1) was higher and the response was more rapid in Arg16 homozygotes than carriers of the Gly16 variant (18% versus 4.9%, respectively; P<0.03).12
Israel et al.,13 performed retrospective genotyping of 190 subjects who had mild asthma and who were randomized to albuterol as-needed or two puffs four times daily. The results showed that Arg16 homozygous asthmatics on regular albuterol for 16 weeks had reduced morning peak expiratory flow rates from baseline (30.5±12.1 L/min) relative to homozygous Arg16 asthmatics on as-needed albuterol (P=0.012). Moreover, homozygous Gly16 asthmatics were not affected by regular treatment. A study of the relationships between β2AR polymorphisms and clinical outcomes showed that homozygous Arg16 asthmatics taking regularly scheduled salbutamol had a higher rate of asthma exacerbation than those taking a placebo.14
A study by Wechsler et al.,15 showed that homozygous Arg16 was possibly associated with a reduced response to LABA as well, and this has led to concerns that the use of combined ICS/LABA medications would result in poor clinical outcomes in patients with polymorphisms of this gene. However, a recent study suggested that the Gly16Arg polymorphism does not predict worse outcomes in moderate-to-severe asthmatic patients treated with LABA/ICS.16 In that study, asthmatic patients were treated with ICS/LABA, and the β2AR polymorphism had no effect on the asthma exacerbations; therefore, it offers reassurance for the continued use of combined ICS/LABA therapy.
Glucocorticoids are the most potent anti-inflammatory drugs used for asthma treatment. Inhaled glucocorticoids are established as the first-line treatment in adults and children with persistent asthma. However, clinical studies have shown that about 5% to 10% of all patients with asthma, and up to 35% of those with severe disease, have reduced responses to glucocorticosteroid therapy.17
Many cases of glucocorticoid resistance may be due to mutations or polymorphisms present in the glucocorticoid receptor (GR) gene (NR3C1).18 Four of these mutations are missense mutations (D641V, G679S, V729I, and I747M), which lead to decreased affinity for the steroid and an impaired ability to activate gene transcription. The N363S polymorphism (rs6195) appears to be associated with an increased sensitivity to glucocorticoids.
Corticotropin releasing hormone receptor 1 (CRHR1) is the primary receptor mediating the release of corticotrophin-releasing hormone, which regulates endogenous cortisol levels and encoded by CRHR1 gene. Genetic variation in CRHR1 is associated with an increased pulmonary function response to inhaled corticosteroids in adults and children.19 Over an 8-week period, the mean FEV1 improvement in adults homozygous for the minor allele (GAT) was 13.7% versus 5.5% in those homozygous for the wild-type allele. Improvement in those heterozygous for the GAT haplotype was intermediate between the two groups, suggesting an additive effect.
More recently, attention has been focused on the possible implication of chromatin remodeling and glucocorticoid resistance in asthma. Histone acetyl transferase (HAT) regulates gene expression through acetylation of core histones, and histone deacetylase (HDAC) mediates gene repression. The level of HAT activity is elevated in patients with asthma, and HDAC activity is reduced in bronchial biopsies, which may contribute to local proinflammatory cytokine production.20 Glucocorticoids switch off inflammatory genes in those with asthma by directly inhibiting HAT activity and by recruiting HDAC2 to activate nuclear factor κB (NF-κB) complexes. Furthermore, dexamethasone-induced repression of Th2 cytokines requires the GR to transport HDAC1 to GATA3 located on the Th2 gene promoters. HDAC6 inactivation leads to hyperacetylation of heat shock protein 90 (Hsp90), a GR chaperone protein, and dissociation from the GR-hsp90 complex results in a reduction in nuclear translocation.21 HDAC10 is a member of the class II HDACs and is found to interact with other HDACs. Known two splice variants of HDAC10 could affect the function of other HDACs.22 In summary, the anti-inflammatory actions of glucocorticoids require the presence or activation of distinct HDACs and HDAC polymorphisms may be associated with glucocorticoid responsiveness.
Antileukotriene therapies inhibit leukotriene synthesis by inhibiting 5-lipoxygenase (5-LO) or by blocking the cysteinyl leukotriene receptor. In a retrospective study assessing the pharmacogenetic significance 5-LO gene (ALOX5) Sp1/Egr1 polymorphism, those possessing only mutant ALOX5 alleles were relatively resistant to treatment with the ALOX5 inhibitor ABT-761. Mean FEV1 improved in wild-type homozygotes (18.8±3.6%) and heterozygotes (23.3±6%) compared to homozygote mutant individuals (-1.2±2.9%).23 Similarly, a significantly higher FEV1 response to the leukotriene receptor antagonist (LTRA) montelukast was observed for the ALOX5 SNP (rs2115819) GG homozygotes compared to those with the AA and AG genotypes (P=0.017).24
Studies on the leukotriene C4 synthase gene (LTC4S) polymorphism (-444 A>C, rs730012) show an improved response to LTRAs in patients carrying the C allele.24,25 Although some studies have failed to reproduce these findings,26-29 perhaps due to different populations and different endpoints, this polymorphism is a good candidate for pharmacogenomics.
A pharmacogenetic study of multi-drug related protein 1 (MRP1), one of the LTC4 transmembrane transporters for the extracellular space, showed that the presence of the T allele (intronic SNP, rs119774) significantly improved FEV1 response to montelukast in asthmatics.24
In a Korean study, the cysteinyl leukotriene receptor 1 gene (CYSLTR1) polymorphism, the minor alleles of both the promoter and coding SNPs [-945 C>T (rs321029) and 927 T>C (rs320995)] were associated with a requirement for montelukast in 89 aspirin-intolerant asthmatics (P<0.02).26 In addition, a study of thromboxane A2 receptor gene (TBXA2R) polymorphism showed that asthmatic children with combinations of the TBXA2R+924T>C homozygote (TT) and TBXA2R+795 T>C hetero- or homozygote (CT or CC) had a 3.67-fold poor FEV1 response to 8-week montelukast treatment than patients with the common alleles.30
Another study on IL-13 polymorphisms revealed a significant association between responsiveness to montelukast and IL-13-1,112 C/T polymorphism and the haplotype of IL-13 polymorphisms.31
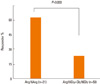
Although the effectiveness of long-acting anticholinergics is well established for managing chronic obstructive pulmonary disease, their roles in asthma management remain controversial. Recently, several studies have suggested the efficacy of anticholinergics in severe asthma,32,33 and interest about the use of long-acting anticholinergics (e.g. tiotropium bromide) for asthma have increased. We published a pharmacogenetic study on tiotropium bromide,34 which included 138 severe asthmatics with decreased lung function who were taking conventional medications, with 18 µg of tiotropium added once daily. Responders were defined as those who improved ≥15% and 200 ml in FEV1 and who maintained the therapy for at least 8 successive weeks. The results showed that 33.3% responded to tiotropium, and Arg16Gly in ADRB2 was significantly associated with the tiotropium response (P=0.003) (Fig. 3). These results suggest that as many as 30% of severe asthmatics benefited from adding tiotropium, and that Arg16Gly in ADRB2 may be a useful marker for tiotropium response.
Pharmacogenomics has the potential to revolutionize asthma therapy. Emerging information and pharmacogenomic study results indicate that standard treatment guidelines cannot be uniformly applied to the entire disease population. Understanding the impact of genetic variations on the therapy response can decrease side effects and improve patient outcomes. Results from a larger number of asthma studies with different populations will be useful for establishing new asthma treatment guidelines and will help realize the ultimate goal of personalized medicine for asthma in the near future.
Figures and Tables
Fig. 2
Long-term agonist-promoted downregulation of the wild-type and mutated β2-adrenergic receptor (modified from Green et al.10).

References
1. Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald M, Gibson P, Ohta K, O'Byrne P, Pedersen SE, Pizzichini E, Sullivan SD, Wenzel SE, Zar HJ. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008. 31:143–178.
2. Lugogo NL, Kraft M. Epidemiology of asthma. Clin Chest Med. 2006. 27:1–15. v.
3. Jee HM, Kim KW, Kim CS, Sohn MH, Shin DC, Kim KE. Prevalence of asthma, rhinitis and eczema in korean children using the International Study Of Asthma and Allergies In Childhood (ISAAC) questionnaires. Pediatr Allergy Respir Dis. 2009. 19:165–172. Korean.
4. Lee SI. Prevalence of childhood asthma in Korea: international study of asthma and allergies in childhood. Allergy Asthma Immunol Res. 2010. 2:61–64.
5. Cho SH, Park HW, Rosenberg DM. The current status of asthma in Korea. J Korean Med Sci. 2006. 21:181–187.
6. Bateman ED, Boushey HA, Bousquet J, Busse WW, Clark TJ, Pauwels RA, Pedersen SE. GOAL Investigators Group. Can guideline-defined asthma control be achieved The Gaining Optimal Asthma ControL study. Am J Respir Crit Care Med. 2004. 170:836–844.
7. Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000. 56:1054–1070.
8. Johnson M. Molecular mechanisms of beta(2)-adrenergic receptor function, response, and regulation. J Allergy Clin Immunol. 2006. 117:18–24. quiz 5.
9. Maxwell TJ, Ameyaw MM, Pritchard S, Thornton N, Folayan G, Githang'a J, Indalo A, Tariq M, Mobarek A, Evans DA, Ofori-Adjei D, Templeton AR, McLeod HL. Beta-2 adrenergic receptor genotypes and haplotypes in different ethnic groups. Int J Mol Med. 2005. 16:573–580.
10. Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994. 33:9414–9419.
11. Cho SH, Oh SY, Bahn JW, Choi JY, Chang YS, Kim YK, Min KU, Kim YY. Association between bronchodilating response to short-acting beta-agonist and non-synonymous single-nucleotide polymorphisms of beta-adrenoceptor gene. Clin Exp Allergy. 2005. 35:1162–1167.
12. Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther. 1999. 65:519–525.
13. Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, Cooper DM, Fahy JV, Fish JE, Ford JG, Kraft M, Kunselman S, Lazarus SC, Lemanske RF, Martin RJ, McLean DE, Peters SP, Silverman EK, Sorkness CA, Szefler SJ, Weiss ST, Yandava CN. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000. 162:75–80.
14. Taylor DR, Drazen JM, Herbison GP, Yandava CN, Hancox RJ, Town GI. Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax. 2000. 55:762–767.
15. Wechsler ME, Lehman E, Lazarus SC, Lemanske RF Jr, Boushey HA, Deykin A, Fahy JV, Sorkness CA, Chinchilli VM, Craig TJ, Di-Mango E, Kraft M, Leone F, Martin RJ, Peters SP, Szefler SJ, Liu W, Israel E. beta-Adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006. 173:519–526.
16. Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007. 370:2118–2125.
17. Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006. 117:522–543.
18. Bray PJ, Cotton RG. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003. 21:557–568.
19. Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, Liggett SB, Gelfand EW, Rosenwasser LJ, Richter B, Israel E, Wechsler M, Gabriel S, Altshuler D, Lander E, Drazen J, Weiss ST. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004. 13:1353–1359.
20. Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, Adcock IM. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med. 2002. 166:392–396.
21. Bhavsar P, Ahmad T, Adcock IM. The role of histone deacetylases in asthma and allergic diseases. J Allergy Clin Immunol. 2008. 121:580–584.
22. Fischer DD, Cai R, Bhatia U, Asselbergs FA, Song C, Terry R, Trogani N, Widmer R, Atadja P, Cohen D. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J Biol Chem. 2002. 277:6656–6666.
23. Drazen JM, Yandava CN, Dube L, Szczerback N, Hippensteel R, Pillari A, Israel E, Schork N, Silverman ES, Katz DA, Drajesk J. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999. 22:168–170.
24. Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, Wang J, Sylvester J, Holbrook J, Wise R, Weiss ST, Barnes K. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006. 173:379–385.
25. Asano K, Shiomi T, Hasegawa N, Nakamura H, Kudo H, Matsuzaki T, Hakuno H, Fukunaga K, Suzuki Y, Kanazawa M, Yamaguchi K. Leukotriene C4 synthase gene A(-444)C polymorphism and clinical response to a CYS-LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma. Pharmacogenetics. 2002. 12:565–570.
26. Kim SH, Ye YM, Hur GY, Lee SK, Sampson AP, Lee HY, Park HS. Cys-LTR1 promoter polymorphism and requirement for leukotriene receptor antagonist in aspirin-intolerant asthma patients. Pharmacogenomics. 2007. 8:1143–1150.
27. Klotsman M, York TP, Pillai SG, Vargas-Irwin C, Sharma SS, van den Oord EJ, Anderson WH. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007. 17:189–196.
28. Currie GP, Lima JJ, Sylvester JE, Lee DK, Cockburn WJ, Lipworth BJ. Leukotriene C4 synthase polymorphisms and responsiveness to leukotriene antagonists in asthma. Br J Clin Pharmacol. 2003. 56:422–426.
29. Lee SY, Kim HB, Kim JH, Kim BS, Kang MJ, Jang SO, Seo HJ, Hong SJ. Responsiveness to montelukast is associated with bronchial hyperresponsiveness and total immunoglobulin E but not polymorphisms in the leukotriene C4 synthase and cysteinyl leukotriene receptor 1 genes in Korean children with exercise-induced asthma (EIA). Clin Exp Allergy. 2007. 37:1487–1493.
30. Kim JH, Lee SY, Kim HB, Jin HS, Yu JH, Kim BJ, Kim BS, Kang MJ, Jang SO, Hong SJ. TBXA2R gene polymorphism and responsiveness to leukotriene receptor antagonist in children with asthma. Clin Exp Allergy. 2008. 38:51–59.
31. Kang MJ, Lee SY, Kim HB, Yu J, Kim BJ, Choi WA, Jang SO, Hong SJ. Association of IL-13 polymorphisms with leukotriene receptor antagonist drug responsiveness in Korean children with exercise-induced bronchoconstriction. Pharmacogenet Genomics. 2008. 18:551–558.
32. Iwamoto H, Yokoyama A, Shiota N, Shoda H, Haruta Y, Hattori N, Kohno N. Tiotropium bromide is effective for severe asthma with noneosinophilic phenotype. Eur Respir J. 2008. 31:1379–1380.
33. Kapoor AS, Olsen SR, O'Hara C, Puttagunta L, Vethanayagam D. The efficacy of tiotropium as a steroid-sparing agent in severe asthma. Can Respir J. 2009. 16:99–101.
34. Park HW, Yang MS, Park CS, Kim TB, Moon HB, Min KU, Kim YY, Cho SH. Additive role of tiotropium in severe asthmatics and Arg-16Gly in ADRB2 as a potential marker to predict response. Allergy. 2009. 64:778–783.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download