Abstract
Allergic diseases such as bronchial asthma and atopic dermatitis develop by a combination of genetic and environmental factors. Several candidate causative genes of asthma and atopy have been reported as the genetic factors. The clinical features of patients and causes of diseases vary. Therefore, personalized medicine (tailor-made medicine) is necessary for the improvement of quality of life (QOL) and for asthma cure. Pharmacogenetics is very important for personalized medicine. Here, we present the genetics and pharmacogenetics of asthma in children. Finally, we show the guideline for personalized medicine for asthma, particularly in childhood, including the pharmacogenetics of anti-asthmatic drugs, preliminarily produced by the authors.
Allergic diseases such as bronchial asthma and atopic dermatitis develop as a result of a combination of genetic and environmental factors. Several candidate causative genes of asthma and atopy have been reported as the genetic factors.
Recently treatment/management guidelines on bronchial asthma and many other disorders have been published, and they are used in clinical practice. However, the clinical features of patients and causes of diseases vary. Therefore, personalized medicine (tailor-made medicine) is necessary for the improvement of quality of life (QOL) and for asthma cure. Pharmacogenetics is very important for personalized medicine.
Here, we present the genetics and pharmacogenetics of asthma in children.
There is sufficient evidence to indicate that asthma is hereditary. A number of studies have shown an increased prevalence of asthma and the phenotype associated with asthma among the offspring of subjects with asthma compared with the offspring of subjects without asthma.
Many studies have shown that there is a genetic accumulation in the development of asthma and allergic disorders. Therefore, the development of asthma and allergic disorders is correlated with some genes. We consider that multiple causative genes are involved, and not a single gene, because there are multiple pathogeneses of asthma and allergic reactions.
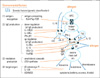
Many candidate genes related to the development of asthma and atopy have been identified, and different genes may be involved in different ethnic groups.1 Among more than 100 genes determined from candidate gene association studies, 79 genes are associated with an asthma- or atopy-related phenotype in 2 or more independent study samples.2
Next, we focus on the several genes related to the development of asthma and atopy, in accordance with the various stages of allergic reaction and development of asthma and atopy.
IgE production is upregulated by Th2 cytokines, particularly interleukin-4 (IL-4), and is downregulated by Th1 cytokines, particularly interferon-γ (IFN-γ). Interleukin-12 (IL-12) and interleukin-18 (IL-18) are the cytokines that induce IFN-γ and downregulate IgE production.5 We review the genetic variation of the cytokine signaling in atopy, and enhanced IgE production.
This item is divided into two parts, (i) "Genes related to upregulation of IgE production in asthma and atopy" and (ii) "Genes related to downregulation of IgE production in asthma and atopy."
Several linkage analyses and mutations of candidate genes of atopy (i.e., enhanced IgE production) have been reported. In 1989, Cookson et al.6 reported a linkage between IgE responses underlying asthma and rhinitis and chromosome 11q. Moreover, Shirakawa et al.7 reported that a common variant of FcεRIβ on chromosome 11, Ile181Leu within the 4th transmembrane domain, shows significant association with positive IgE responses. Several associations have been noted between atopy and genes on the chromosome 5 cytokine cluster, including IL-4.
An Ile50Val (numbering for mature peptide) variant of human IL-4Rα has been identified. In 1998, Mitsuyasu et al.8 reported that the Ile50Val variant of the IL-4Rα chain upregulates IgE synthesis and is associated with atopic asthma. Ile50 is associated with atopic asthma but not with nonatopic asthma; Ile50 is specifically and significantly associated with increased total serum IgE levels and mite-specific IgE. The association with atopy was particulary strong in children.8 The data from both mouse and human cell lines strongly suggest that the Ile50 variant of IL-4Rα significantly upregulates receptor response to IL-4, with a resultant increased activation of Stat6, and hence, increased cell proliferation and increased IgE production. Moreover, Shirakawa et al.9 noted genetic variants of IL-13.
The genetic defects in the downregulation (brake) of IgE production, particularly in terms of IL-12 and IL-18 signalings, are discussed. We found that the reduced IFN-γ production by peripheral blood mononuclear cells (PBMCs) following stimulation with IL-12 or IL-18 is associated with heterozygous IL-12 receptor β2 (IL-12Rβ2) chain gene mutations (2496 del 91, 1577 A to G (Arg 313 Gly), 2799 A to G (His 720 Arg) or IL-18 receptor α (IL-18Rα) chain gene mutation (del 950 CAG) in atopic subjects.10,11
We identified a novel heterozygous single-nucleotide substitution 1400 T to C (Leu 467 Pro), in the seventh exon of the IFN-γ receptor 1 (IFN-γR1) chain gene.12 This substitution was detected in six of 89 allergic patients (including asthma), but not in 72 nonallergic subjects. There was a difference in the Leu 467 Pro frequency between allergic and nonallergic subjects (P<0.05). The serum IgE levels of the allergic patients with the Leu 467 Pro substitution were higher than those of the nonallergic subjects (P<0.001). These results suggest that Leu 467 Pro in the IFN-γR1 chain gene is one of the candidate susceptibility genes for asthma or atopic diseases.
The locus of leukotriene C4 synthase (LTC4S) is on chromosome 5q35 and has been associated with allergic diseases on the basis of a genomewide search. Cysteiny1 leukotrienes (cysLTs) play important roles in asthma and can mediate bronchial smooth muscle constriction and increase mucous secretion, vascular permeability, and cellular infiltration.13 LTC4S converts LTA4 to LTC4 by conjugation to reduced glutathione. A single-nucleotide promoter polymorphism (A-444C) in LTC4S has been associated with aspirin-sensitive asthma,14,15 although recent studies have found no association between this promoter polymorphism and aspirin-sensitive asthma.16
Very recently, we have reported that a novel single-nucleotide substitution 10G>A (Glu 4 Lys) in LTC4S is associated with asthma.17
Nitric oxide (NO) is produced by a group of enzymes referred to as nitric oxide synthase: endothelial (eNOS), neuronal (nNOS), and inducible NOS (iNOS). The association of some nNOS markers with asthma or related phenotypes has been reported.18
There was no relationship between β2-adrenergic receptor (ADRβ2) polymorphisms and asthma prevalence, but the Gly-16 variant was apparently associated with a more severe form of asthma.19 Subsequently, Turki et al.20 found that the Gly-16 allele is more frequent among subjects with nocturnal asthma than among nonnocturnal asthmatics.
Van Eerdewegh and Holgate et al.21 performed a genomewide scan on 460 Caucasian families and identified a locus on chromosome 20p13 that is linked to asthma (Log10 of the likelihood ratio LOD, 2.94) and bronchial hyperresponsiveness (LOD, 3.93). A survey of 135 polymorphisms in 23 genes identified the ADAM33 gene as being significantly associated with asthma using case control, transmission disequilibrium and haplotype analyses (P=0.04-0.000003). ADAM proteins are membrane-anchored metalloproteases with diverse functions, which include the shedding of cell-surface proteins such as cytokines and cytokine receptors. The identification and characterization of ADAM33, a putative asthma susceptibility gene identified by positional cloning in an outbred population, should provide insights into the pathogenesis and natural history of this common disease.
On the basis of many reports and our results, we present a new genetic classification of atopy and asthma in Fig. 1.22 There are four categories of genes that control the expression of allergic disorders, which include (i) antigen recognition, (ii) IgE production (downregulation=brake, and upregulation), (iii) production and release of mediators, and (iv) events on target organs. This genetic classification will facilitate the development of pharmacogenetics and personalized medicine (tailor-made medicine).
The response of an individual patient to any given drug depends on several factors such as pathogenesis of disease, compliance, disease severity, and genetic background.23 The great hope in pharmacogenetics is that it will help predict either treatment response (efficacy) or the risk of adverse drug reactions in the general population and that it will prove cost-effective to genotype individuals before treatment,23 that is, for personalized medicine. Here, we review the pharmacogenetics of anti-asthmatic drugs.
Beta2-agonists act via binding to the β2-adrenergic receptor (ADRβ2), that is, a cell surface G-protein-coupled receptor. The β2-adrenergic receptor has several polymorphisms in the coding region, such as Arg 16 Gly, Gln 27 Glu, and Thr 164 ILe. The Arg 16 Gly and Gln 27 Glu are functionally important. Patients with homozygous for Arg 16 did worse clinically in terms of the major end points and showed adverse effects, rather than homozygous for Gly 16.24-26
Leukotrienes are very important mediators in asthma in children as well as in adults. Leukotrienes are released by eosinophils, mast cells, and alveolar macrophages. In the production of leukotrienes, several enzymes such as 5-lipoxygenase (ALOX5) and leukotriene C4 synthase (LTC4S) are important. LTC4S is a membrane-bound glutathione transferase, and synthesizes cysteinyl-leukotrienes, converting LTA4 to LTC4.
In the treatment with zafirlukast (one of the leukotriene receptor antagonists), patients with the homozygous for A-444 showed a lower FEV1 response than those with the A/C or C/C genotype.27
Suplatast tosilate is a Th2 cytokine inhibitor.30 In our recent study, treatment with suplatast tosilate was more effective in children without the -444 A/C polymorphism of the LTC4S gene, and in children without the IL-13 variant Arg 110 Gln. In children who responded well, IFN-γ production was significantly increased after treatment.
Inhaled corticosteroids (ICS) are very effective for asthma in children as well as adults. However, there are patients with severe asthma who show no response to treatment with ICS. There are reports concerning the pharmacogenetics of corticosteroids. The IL-4 589T allele (589 C/T SNP) was found to be associated with ICS-resistant asthma.31,32 The TBX21 (T-bet) gene (His 33 Gln) and the corticotropin-releasing hormore receptor 1 (CRHR1) gene are important for the pharmacogenetics of ICS.33 Particularly in Caucasian children, the heterozygous TBX21 33 His/Gln individuals demonstrated a marked improvement in airway hyperresponsiveness compared with 33 His/His homozyotes on ICS therapy.33,34
The clinical features of patients and causes of diseases vary. Therefore, personalized medicine is necessary for the improvement of QOL and for asthma cure. Pharmacogenetics is very important for personalized medicine.
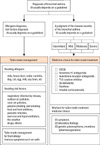
Here, we show the guideline for personalized medicine (tailor-made medicine) for asthma, particularly in children, including the pharmacogenetics of anti-asthmatic drugs, preliminarily produced by the authors (Figs. 2, 3).
Asthma develops by a combination of genetic and environmental factors. There are many environmental factors such as allergens (house dust, mites, foods, and pets so on), viral infections, exercise, passive smoking, and air pollution. The environmental factors are different for each patient. Therefore, it is very important to first eliminate each environmental factor for each patient and to take preventive measures against each environmental factor (Fig. 2), that is, the personalized preventive measures. The preliminary guideline for personalized medicine (tailor-made medicine) for asthma has been published, on the basis of various clinical symptoms, various laboratory findings, as well as pharmacogenetics of anti-asthmatic drugs.
Figure 3 shows the guideline for personalized medicine. For example, as one of the markers of symptoms, for viral-induced asthma, leukotriene receptor antagonists are effective. For exercise induced asthma, disodium cromoglycate (DSCG), leukotriene receptor antagonists, or β2-agonists are effective. As one of the laboratory findings, in the patients with poor IFN-γ production, Th2 cytokine inhibitor is effective. As one of the gene markers, in LTC4S-444 A/A type, Th2 cytokine inhibitor is effective, and in LTC4S-444 A/C or C/C type, leukotriene receptor antagonists are effective. In TBX21 (T-bet) 33 His/Gln or Gln/Gln, ICS is effective.
In the near future, the guidelines for the personalized medicine (tailor-made medicine) for asthma will become more available. For personalized medicine, it is necessary for us to obtain both the approval of an ethical committee and informed consent.
Figures and Tables
Fig. 3
Medicine choice for personalized medicine based on the symptoms, laboratory findings and pharmacogenetics.
○: positive markers; (○): possible markers.
DSCG, disodium cromoglycate; H1-antagonists, histamine H1-receptor antagonists; LTRA, leukotriene receptor antagonists; ICS, inhaled corticosteroids; LT, leukotriene; LTC4S, leukotriene C4 synthase; ALOX5, 5-lipoxygenase; MRP1, Multidrug resistance-associated protein 1; ADRβ2, β2-aderenergic receptor; CRHR1, corticotropin-receptor 1 releasing hormone; R, Arginine; G, Glycine; H, Histidine; Q, Glutamine.

ACKNOWLEDGMENTS
This study was supported by Health and Labour Science Research Grants from the Ministry of Health, Labour and Welfare for Research on Allergic Disease and Immunology.
References
1. Global Initiative for Asthma (GINA): Global Strategy for Asthma Management and Prevention. 2006. Available from: http://www.ginasthma.org.
2. Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes and Immunity. 2006. 7:95–100.
3. Juhn YJ, Kita H, Lee LA, Smith RW, Bagniewski SM, Weaver AL, Pankratz VS, Jocobson RM, Poland GA. Childhood asthma and human leukocyte antigen type. Tissue Antigens. 2007. 69:38–46.
4. Juhn YJ, Kita H, Bagniewski SM, Weaver AL, Pankratz VS, Jocobson RM, Poland GA. Severity of childhood asthma and human leukocyte antigens type. J Asthma. 2007. 44:163–168.
5. Kondo N, Matsui E, Kaneko H, Fukao T, Teramoto T, Kato Z, Onishi H, Nishimura A. Pawankar R, Holgate ST, Rosenwasser LJ, editors. Genetics of pediatric asthma. Allergy frontiers volume 1 allergy frontiers: epigenetics, allergens and risk factors. 2009. Heidelberg: Springer;189–203.
6. Cookson WO, Sharp PA, Faux JA, Hopkin JM. Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet. 1989. 1:1292–1295.
7. Shirakawa T, Li A, Dubowitz M, Dekker JW, Shaw AE, Faux JA, Ra C, Cookson WO, Hopkin JM. Association between atopy and variants of the beta subunit of the high-affinity immunoglobulin E receptor. Nat Genet. 1994. 7:125–130.
8. Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin JM. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998. 19:119–120.
9. Shirakawa T, Deichmann KA, Izuhara I, Mao I, Adra CN, Hopkin JM. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol Today. 2000. 21:60–64.
10. Matsui E, Kaneko H, Fukao T, Teramoto T, Inoue R, Watanabe M, Kasahara K, Kondo N. Mutations of the IL-12 receptor β2 chain gene in atopic subjects. Biochem Biophy Res Commun. 1999. 266:551–555.
11. Watanabe M, Kaneko H, Shikano H, Aoki M, Sakaguchi H, Matsui E, Inoue R, Kato Z, Kasahara K, Fukutomi O, Kondo T, Kondo N. Predominant expression of 950delCAG of IL-18R alpha chain cDNA is associated with reduced IFN-gamma production and high serum IgE levels in atopic Japanese children. J Allergy Clin Immunol. 2002. 109:669–675.
12. Aoki M, Matsui E, Kaneko H, Inoue R, Fukao T, Watanabe M, Teramoto T, Kato Z, Suzuki K, Suzuki Y, Kasahara K, Kondo N. A novel single-nucleotide substitution, Leu 467 Pro, in the interferon-gamma receptor 1 gene associated with allergic diseases. Int J Mol Med. 2003. 12:185–191.
13. Henderson WR Jr. The role of leukotrienes in inflammation. Ann Intern Med. 1994. 121:684–697.
14. Sanak M, Hu S, Szczeklik A. Leukotriene C4 synthase promoter polymorphism and risk of aspirin-induced asthma. Lancet. 1997. 29:1599–1600.
15. Kim SH, Hur GY, Choi JH, Park HS. Pharmacogenetics of aspirin-intolerant asthma. Pharmacogenomics. 2008. 9:85–91.
16. Van Sambeek R, Stevenson DD, Baldasaro M, Lam BK, Zhao J, Yoshida S, Yandora C, Drazen JM, Penrose JF. 5'flanking region polymorphism of the gene encoding leukotriene C4 synthase does not correlate with the aspirin-intolerant asthma phenotype in the United States. J Allergy Clin Immunol. 2000. 106:72–76.
17. Yoshikawa K, Matsui E, Kaneko H, Fukao T, Inoue R, Teramoto T, Shinoda S, Fukutomi O, Aoki M, Kasahara K, Kondo N. A novel single-nucleotide substitution, Glu 4 Lys, in the leukotriene C4 synthase gene associated with allergic diseases. Int J Mol Med. 2005. 16:827–831.
18. Gao PS, Kawada H, Kasamatsu T, Mao XQ, Roberts MH, Miyamoto Y, Yoshimura M, Saitoh Y, Yasue H, Nakao K, Adra CN, Kun JF, Moro-oka S, Inoko H, Ho LP, Shirakawa T, Hopkin JM. Variants of NOS1, NOS2, and NOS3 genes in asthmatics. Biochem Biophys Res Commun. 2000. 267:761–763.
19. Reihsaus EM, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol. 1993. 8:334–339.
20. Turki J, Pak J, Green SA, Martin RJ, Liggett SB. Genetic polymorphisms of the beta 2-adrenergic receptor in nocturnal and non-nocturnal asthma. Evidence that Gly16 correlates with the nocturnal phenotype. J Clin Invest. 1995. 95:1635–1641.
21. Van Eerdewegh P, Little RD, Dupuis J, Mastro RGD, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002. 418:426–430.
22. Kondo N, Matsui E, Kaneko H, Kato Z, Fukao T, Teramoto T, Shikano H, Aoki M, Onishi H, Tatebayashi K, Omoya K, Kondo M, Matsukuma E, Kasahara K, Morimoto N. Genetic defects in downregulation of IgE production and a new genetic classification of atopy. Allergology International. 2004. 53:77–85.
23. Hall IP. Pharmacogenetics of asthma. Chest. 2006. 130:1873–1878.
24. Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, Cooper DM, Fahy JV, Fish JE, Ford JG, Kraft M, Kunselman S, Lazarus SC, Lemanske RF, Martin RJ, McLean DE, Peters SP, Silverman EK, Sorkness CA, Szefler SJ, Weiss ST, Yandava CN. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000. 162:75–80.
25. Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, Deykin A, Fagan JK, Fahy JV, Fish J, Kraft M, Kunselman SJ, Lazarus SC, Lemanske RF Jr, Liggett SB, Martin RJ, Mitra N, Peters SP, Silverman E, Sorkness CA, Szefler SJ, Wechsler ME, Weiss ST, Drazen JM; National Heart, Lung, and Blood Institute's Asthma Clinical Research Network. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004. 364:1505–1512.
26. Wechsler ME, Lehman E, Lazarus SC, Lemanske RF Jr, Boushey HA, Deykin A, Fahy JV, Sorkness CA, Chinchilli VM, Craig TJ, DiMango E, Kraft M, Leone F, Martin RJ, Peters SP, Szefler SJ, Liu W, Israel E; National Heart, Lung, and Blood Institute's Asthma Clinical Research Network. beta-Adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006. 173:519–526.
27. Palmer LJ, Silverman ES, Weiss ST, Drazen JM. Pharmacogenetics of asthma. Am J Respir Crit Care Med. 2002. 165:861–866.
28. Sampson AP, Siddiqui S, Buchanan D, Howarth PH, Holgate ST, Holloway JW, Sayers I. Variant LTC(4) synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax. 2000. 55:Suppl 2. S28–S31.
29. Asano K, Shiomi T, Hasegawa N, Nakamura H, Kudo H, Matsuzaki T, Hakuno H, Fukunaga K, Suzuki Y, Kanazawa M, Yamaguchi K. Leukotriene C4 synthase gene A(-444)C polymorphism and clinical response to a CYS-LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma. Pharmacogenetics. 2002. 12:565–570.
30. Tamaoki J, Kondo M, Sakai N, Aoshiba K, Tagaya E, Nakata J, Isono K, Nagai A. Effect of suplatast tosilate, a Th2 cytokine inhibitor, on steroid-dependent asthma: a double-blind randomised study. Lancet. 2000. 356:273–278.
31. Leung DY, Bloom JW. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2003. 111:3–22.
32. Tantisira KG, Weiss ST. The pharmacogenetics of asthma: an update. Curr Opin Mol Ther. 2005. 7:209–217.
33. Szalai C, Ungvári I, Pelyhe L, Tölgyesi G, Falus A. Asthma from a pharmacogenomic point of view. Br J Pharmacol. 2008. 153:1602–1614.
34. Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, Peng SL, Drazen JM, Glimcher LH, Weiss ST. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci USA. 2004. 101:18099–18104.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download