Abstract
BACKGROUND/OBJECTIVES
Feeding in infancy is the most significant determinant of the intestinal microbiota in early life. The aim of this study was to determine the gut microbiota of Korean infants and compare the microbiota obtained between breast-fed and formula-fed Korean infants.
SUBJECTS/METHODS
We analyzed the microbial communities in fecal samples collected from twenty 4-week old Korean (ten samples in each breast-fed or formula-fed) infants using pyrosequencing.
RESULTS
The fecal microbiota of the 4-week-old Korean infants consisted of the three phyla Actinobacteria, Firmicutes, and Proteobacteria. In addition, five species, including Bifidocbacterium longum, Streptococcus salivarius, Strepotococcus lactarius, Streptococcus pseudopneumoniae, and Lactobacillus gasseri were common commensal intestinal microbiota in all infants. The predominant intestinal microbiota in the breast-fed infants (BFI) included the phylum Actinobacteria (average 70.55%), family Bifidobacteriacea (70.12%), genus Bifidobacterium (70.03%) and species Bifidobacterium longum (69.96%). In the microbiota from the formula-fed infants (FFI), the proportion of the phylum Actinobacteria (40.68%) was less, whereas the proportions of Firmicutes (45.38%) and Proteobacteria (13.85%) as well as the diversity of each taxonomic level were greater, compared to those of the BFI. The probiotic species found in the 4-week-old Korean infants were Bifidobacterium longum, Streptococcus salivarius, and Lactobacillus gasseri. These probiotic species accounted for 93.81% of the microbiota from the BFI, while only 63.80% of the microbiota from the FFI. In particular, B. longum was more abundant in BFI (69.96%) than in FFI (34.17%).
CONCLUSIONS
Breast milk supports the growth of B. longum and inhibits others. To the best of our knowledge, this study was the first attempt to analyze the gut microbiota of healthy Korean infants according to the feeding type using pyrosequencing. Our data can be used as a basis for further studies to investigate the development of intestinal microbiota with aging and disease status.
The human intestinal microbiota is a complex and dynamic ecosystem with 300-500 different species of bacteria [1] and plays an important role in maintaining host health since it is involved in nutrition, pathogenesis, and immunology [2].
The gut microbiota of an individual is influenced by different dietary habits during life. The gut microbiota of children fed a modern western diet compared to a rural diet has been characterized [3], and the effect of dietary polyphenols on the human gut microbiota has been reported [4]. Arumugam et al. have proposed that human gut microbial communities represent three predominant variants, "enterotypes," dominated by Bacteroides, Prevotella, and Ruminococcus, respectively [5]. In addition, the enterotype is related to disease states, and long-term dietary interventions may allow modulation of an individual's microbiota, which subsequently improves health [6].
The neonatal period is a crucial stage for gastrointestinal colonization [7], which influences the adult intestinal microbiota and lifelong health [8]. In general, the fetus is known to be sterile, and gut colonization in newborn infants starts rapidly within a few hours by contact with the maternal vagina and the environment [9]. The critical stage of gut colonization occurs within the first few weeks after birth and is influenced by a variety of factors including the environment, diet, gestational age, hospitalization, antibiotics, delivery mode, and feeding method [10]. In early life, one of the most significant determinants of the intestinal microbiota is the type of feeding.
Breast milk is the gold standard for growing infants, because it is a complex species-specific biological fluid adapted to satisfy the nutritional requirements and develop the immune system [11]. In addition, breast milk has been reported to protect against different infectious diseases [12,13,14]. Colostrum and mature milk contain potential antimicrobial factors such as immunoglobulins, cytokines, oligosaccharides, lysozyme, lactoferrin, other glycoproteins, and antimicrobial peptides that can inactivate pathogens individually or synergistically [15].
Recently barcoded pyrosequencing based on the small subunit ribosomal RNA (16S rRNA) gene has been used to analyze the entire intestinal microbiota. This powerful technique provides information regarding the complex microbiota, which had not been previously detected by culture methods [7,16,17]. In this study, we compared the gut microbiota of Korean infants fed breast milk versus formula milk by using pyrosequencing. This pilot study will allow us to better understand the relationship between the gut microbiota and feeding type, and more importantly, to Korean infant health.
Fecal samples were collected from twenty healthy newborn infants at 4 weeks of age who were delivered from healthy mothers by normal spontaneous vaginal delivery at Arante Women's Hospital in Seoul, Korea, from May 2012 to February 2013. Ten infants were exclusively breast-fed, and the other ten infants were formula-fed (some exposure to breast milk but fed 70-100% with formula milk). Meconium and fecal samples were collected from the infants, which were defecated spontaneously on disposable diapers at 4 weeks after birth. The specimens were immediately sealed in a plastic bag and were stored at -80℃. The fecal samples from infants who were delivered by Caesarean section, premature, or give antibiotic prophylaxis or therapy as well as those whose mothers had infections during pregnancy, clinical illness, antibiotic administration, or probiotic supplementation were excluded from this study. This study was approved by the Institutional Review Board of Ewha Womans University (IRB No. 2012-3-10; Seoul, Korea), and all of the participants provided informed consent.
Metagenomic DNA was isolated from fecal samples of infants using a Fast DNA SPIN Kit (MP BIO, Santa Ana, CA, USA). Extraction was performed according to the manufacturer's instructions. DNA was eluted in 50 µl of elution buffer and stored at -20℃ prior to use. The genomic DNA concentration was determined by the optimal density at 260 nm, and the purity was assessed by the ratio of the absorbance at 260 nm and 280 nm.
The PCR mixture was prepared with Red-Taq Ready Mix (Sigma-Aldrich, St. Louis, MO), 40 µM primers (COSMO Genetech, Korea), and 100 ng of genomic DNA in a total volume of 25 µl. For PCR, the primer set 27F and 518R was used to amplify the 27-518 region of the bacterial 16S rRNA containing the V1 and V3 regions [18].
The PCR products were purified with the QIAquick PCR purification kit (Qiagen), and quantified using the PicoGreen dsDNA Assay kit (Invitrogen, Carlsbad, CA). Equimolar concentration of each amplicon were mixed and sequenced on a Roche/454 GS Junior system at ChunLab, Inc (Seoul, Korea).
The sequences obtained from the pyrosequencer were analyzed according to previously described methods [19]. Briefly, the sequences of each sample were sorted by a unique barcode, and low quality reads (average quality score < 25 or read length < 300bp) were removed for further analysis. Primer sequences were trimmed by pairwise alignment, and trimmed sequences were clustered to correct for sequencing errors. Representative sequences in each cluster were identified using the EzTaxon-e database [20]. Chimeric sequences were removed using the UCHIME program [21] and the diversity indices were calculated using the Mothur program [22]. The pyrosequences obtained in this study are available in the EMBL SRA database under the study PRJEB6615 (http://www.ebi.ac.uk/ena/data/view/PRJEB6615).
The CLcommunity™ program was used to analyze the pyrosequencing sequences of the intestinal microbiota. The operational taxonomic unit (OTU) refers to a group of sequences that are mathematically defined with a sequence similarity of 97%. This diversity measure was carried out using CD-HIT methods. Alpha-diversity indices such as ACE, Chao1, and Shannon diversity were used to estimate species richness. All data were expressed as mean ± standard error of the mean (SEM) for each group. Comparison between the two groups of infants was performed with the Wilcoxon-Mann-Whitney test. All statistical analyses were performed with PASW Statistics 18.0 (SPSS Inc., Chicago, IL, USA). P-values < 0.05 were considered statistically significant.
There were no significant differences in terms of the characteristics between the test groups of breast-fed infants (BFI) and formula-fed infants (FFI) (Table 1). The mean maternal age was 31.4 ± 3.3 years old in the BFI group and 32.3 ± 2.3 years old in the FFI group. There were more females than males in the BFI group, whereas there were equal numbers of females and males in the FFI group. The mean birth weights were 3.1 ± 0.4 kg and 3.3 ± 0.5 kg, respectively. All infants were 4 weeks of age. No infants received any additional foods and no mothers took antibiotics after the first week of delivery.
A total of 207,596 sequences were used for the final analysis (Table 2). The average numbers of sequences were 11,100 from the BFI and 9,600 from the FFI, indicating no significant difference (P > 0.05). The Good's coverages of all samples were greater than 0.98, and the total number of OTUs obtained was 737 (average, 73.7 per sample) in the BFI samples and 1,364 (average, 136.4 per sample) in the FFI samples (Table 3). The Shannon diversity indices in the BFI ranged from 0.82 to 1.91, while those in the FFI were from 1.56 to 2.21. These diversity indices indicated that the intestinal microbiota obtained from the FFI was more diverse that from the BFI.
The analyzed sequences of microbiota were classified from the species up to the phylum level. The microbiota of the infant fecal samples consisted of the three phyla Actinobacteria, Firmicutes, and Proteobacteria (Table 4 & Fig. 1). Actinobacteria (average 70.55% of total reads) was the predominant phylum in the BFI, followed by Firmicutes (26.15%) and Proteobacteria (3.29%). In contrast, in the FFI, the proportions of Actinobacteria (40.68%) and Firmicutes (45.38%) were similar, followed by Proteobacteria (13.85%). The relative abundance of Firmicutes and Proteobacteria was higher in the FFI than in the BFI.
Analysis at the family level revealed that the microbiota from the FFI was more diverse than that from the BFI. Bifidobacteriaceae was the most predominant family within the phylum Actinobacteria (Table 4 & Fig. 1), and comprised 70.12% of the BFI samples, compared to 36.61% of the FFI samples. In Actinobacteria from the FFI, Actinomycetaceae (3.73%) was the second most predominant family after Bifidobacteriaceae. The phylum Firmicutes from the BFI consisted mainly of Streptococcaceae (17.31%) and Lactobacillaceae (8.02%), while the five families Streptococcaceae (28.86%), Veillonellaceae (6.12%), Enterococcaceae (4.96%), Lactobacillaceae (2.56%), and Coprobacillus (1.94%) were abundantly detected within Firmicutes from the FFI. Within the phylum Proteobacteria, Enterobacteriaceae was the major family in both the BFI and FFI samples. The proportion of Enterobacteriaceae from the FFI (12.64%) was higher than that from the BFI (3.17%).
The fecal microbiota from individual BFI and FFI was analyzed to the genus level (Tables 5 & 6). Bifidobacterium was remarkably abundant in the BFI, with an average percentage of 70.03%. In BFI 7, 8, 9, and 10, the percentage of Bifidobacterium was below the average amount; instead, the genera Streptococcus and Lactobacillus were more abundant, indicating that total amount of probiotics was stable (Table 5). The main genera in the FFI were Bifidobacterium (average, 35.99%) and Streptococcus (average, 23.10%); however, their percentages were quite varied in each individual (Table 6). The proportion of Bifidobacterium was lower than in the FFI than in the BFI. In addition, the microbiota of the FFI contained a significant amount of the genera Escherichia, Veillonella, Enterococcus, and Enterobacter, whereas the content of Lactobacillus was low.
The species of fecal microbiota in individual infants were compared by heatmap analysis (Fig. 2). The species compositions from the FFI were more diverse than those from the BFI. The five species Bifidocbacterium longum, Streptococcus salivarius, Strepotococcus lactarius, Streptococcus pseudopneumoniae, and Lactobacillus gasseri were found in all infants; therefore, these five species are common commensal intestinal microbiota in 4-week-old Korean infants, independently of feeding type. In addition to these five species, Escherichia coli, Enterococcus faecalis, and Veillonella atypical were abundant in most of the FFI microbiota.
Probiotics are defined as living microorganisms that confer a health benefit on the host when consumed [23]. S. salivarius, L. gasseri, and species of Bifidobacterium were determined to be the dominant probiotics from the infant microbiota in this study. Comparison of probiotics between the BFI and FFI showed that the probiotics in the BFI samples (93.8%) were more abundant than the FFI samples (65.6%) (Fig. 3). The proportion of B. longum was higher in the BFI (70.0%) than in the FFI (34.2%), and the proportion of L. gasseri was also higher in the BFI (7.9%) than in the FFI (2.5%). On the other hand, the proportion of S. salivarius from the FFI (27.2%) was higher than that from the BFI (16.0%). Diverse species of Bifidobacteria such as B. breve, B. bifidum, L. brevis, and L. reuteri were detected only in the FFI. The relative abundance of the genus Bifidobacterium showed a significant difference between the BFI (average 70.0%) and the FFI (average 36.4%) (Wilcoxon-Mann-Whitney test, P = 0.0015) (Fig. 4).
Overall, the dominant fecal bacteria from the 4-week-old Korean infants studied were the probiotics Bifidobacterium longum, Streptococcus salivarius, and Lactobacillus gasseri. In the BFI, probiotics consisted of 93.81% of the total microbiota, of which B. longum was the most predominant species.
In the past few years, determination of the composition of infant intestinal microbiota has made progress through the use of pyrosequencing-based monitoring [24]. Accordingly, the role of the infant intestinal microbiota has been emphasized not only for intestinal health but also lifelong health in adulthood. The present study showed that the fecal microbiota of 4-week-old Korean infants consisted of the three phyla Actinobacteria, Firmicutes, and Proteobacteria in both the BFI and FFI groups. Staphylococcus epidermis was also detected in the BFI, which apparently originated from the mother's skin during breast-feeding [25]. In the microbiota from the BFI, the phylum Actinobacteria (70.55%), family Bifidobacteriacea (70.12%), genus Bifidobacterium (70.03%), and species Bifidobacterium longum (69.96%) were predominant. In the microbiota from the FFI, the proportion of Actinobacteria (40.68%) was less, whereas the proportion of Firmicutes (45.38%) and Proteobacteria (13.85%) as well as the diversity of each taxonomic level were greater than in the BFI. These results demonstrated a specific gut microbiota community for Korean 4-week-old infants that is different from 6-week-old European infants [7], in which the genera Bifidobacterium (40.0%), Bacteroides (11.4%) and enterobacteria (7.5%) were detected. Fallani et al. (2010) have described that there is a geographic difference in the proportion of Bacteroides, Bifidobacteria, and Enterobacteria: higher proportions of Bifidobacteria were found in 6-week-old infants from Northern European countries, whereas a more diverse microbiota with a higher proportion of Bacteroides was found in infants from Southern European countries [7]. They found that Bifidobacteria was dominant in BFI, whereas FFI harbored higher proportions of Bacteroides, Clostridium, and Lactobacillus. In addition, Chinese infants have Firmicutes and Proteobacteria as dominant microbiota in all BFI, FFI, and mixed-fed infants aged 1 to 6 months [26]. They found a significant depletion of Bacteroides and Actinobacteria in the FFI, whereas Bifidobacteriaceae comprised only 8.16% of the microbiota in the BFI. The differences between these results and our results may be explained by the different geographical origins, feeding types, delivery modes, and ages of the infants.
The probiotic species found in this study were Bifidobacterium longum, Streptococcus salivarius, and Lactobacillus gasseri. The sum of the probiotic species comprised 93.81% of the microbiota from the BFI, while only 63.80% of that from the FFI. In particular, B. longum was significantly more abundant in the BFI (69.96%) compared with the FFI (34.17%). The FFI exhibited a more diverse microbiota community composed of various bacterial species compared to the BFI. These results are consistent with previous studies showing that the dominant bacteria in BFI are Bifidobacteriaceae and Lactobacillaceae; however, the FFI had multiple intestinal microbiota composed of Bifidobacteria, Bacteroides, Clostridia, Enterococcus, and Staphylococcus [27,28,29]. The increase in the microbiota diversity of the FFI probably depends on the contents of the formula milk; therefore, further studies are required to investigate the composition of formula milk.
The predominance of Bifidobacteriaceae due to the high proportion of B. longum in the BFI is likely related to the higher diversity of microbiota in the FFI. Breast milk contains a low buffering capacity, casein, calcium, and phosphate, but a high content of lactose, which seem to favor the growth of Bifidobacterium compared to other bacteria [30]. Breast milk differs from cow milks as it contains sialylated and fucosylated oligosaccharides, which are utilized for growth of Bifidobacteria [31,32]. The colonization of Lactobacilli and Bifidobacteria in the intestine inhibits the growth of pathogenic microorganisms, such as Staphylococcus aureus, Salmonella typhimurium, Yersinia enterocolitica, and Clostridium perfringens, by nutrient competition and preventing the adhesion of pathogens [33]. Therefore, the relationship between various contents of formula with the infant intestinal microbiota should be studied in detail.
The bacterial microbiota formed in infants depends on the diet of the infants [34], but the environment during birth, prematurity, and hygiene measures also influence the intestinal microbiota community in early infancy [10]. To minimize the effects of factors other than feeding, we only included subjects who did not take antibiotics after the first week of birth and were delivered via normal spontaneous vaginal delivery. In addition, all subjects were full-term infants.
There were several limitations in this study. Only a relatively small number of samples were analyzed, since pyrosequencing requires a high cost. In addition, samples were analyzed only at a single time point. Meconium was also analyzed, but there were not enough bacteria for DNA extraction. Lastly, some FFI were exposed to breast milk during the 4 weeks of life, and the formula compositions were not investigated for the FFI group.
Maternal dietary supplementation, e.g., with probiotics, may provide a health benefit to infants via improvement of the maternal gastrointestinal tract [35]. However, little is known about the microbiota of infants breast fed by mothers with different diets. Furthermore, infant formula resembling breast milk or the direct addition of Bifidobacterium to infant formula leads to a gut microbiota of FFI similar to that of BFI [36]. Therefore, studying the influence of the mother's dietary behaviors and types of formula on the infant's intestinal microbiota for a longer duration is required.
In conclusion, the present study was the first attempt to analyze the gut microbiota in healthy Korean 4-week-old infants by using pyrosequencing. Furthermore, this study provides a basis for a more comprehensive understanding of the diversity of the intestinal microbiota and valuable information for future studies on the role of the intestinal microbiota in lifelong health.
Figures and Tables
Fig. 1
Aggregate microbiota composition at pylum/family level from fecal samples of 4 wks old infants.
(A) Breast-fed infants, (B) Formula-fed infants.

Fig. 2
Heatmap analysis of the species detected from pyrosequencing of fecal samples from 4 wks old BFI and FFI.
The data contained in the matrix are represented by black and white colors. The minimum ratio of a taxon is displayed as an individual sample. The cut-off value was set to 1%. BFI, Breast-fed infants; FFI, Formula-fed infants

Fig. 3
The compositions of probiotics at the species level in fecal samples from 4-week-old BFI and FFI.
etc., bacteria other than probiotics; BFI, Breast-fed infants; FFI, Formula-fed infants
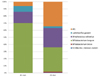
Fig. 4
Scatter plots showing the proportions of Bifidobacterium sequences with respect to the total sequences obtained from pyrosequencing 16S rRNA genes associated with feeding type.
Mean values (horizontal lines) and significance values (P, Mann-Whitney) are shown. BFI, Breast-fed infants; FFI, Formula-fed infants

Table 3
The operational taxonomic units (OTUs) and Shannon diversity index of the fecal microbiota of 4 wks old BFI and FFI

References
1. Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984; 86:174–193.

2. Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012; 28:63–69.

3. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010; 107:14691–14696.

4. Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. 2013; 24:1415–1422.

5. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. MetaHIT Consortium. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180.

6. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334:105–108.

7. Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Doré J. Other Members of the INFABIO Team. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breastfeeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010; 51:77–84.

8. Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr. 2008; 138:1791S–1795S.

9. Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004; 12:129–134.

10. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006; 118:511–521.

11. Hanson LA, Korotkova M. The role of breastfeeding in prevention of neonatal infection. Semin Neonatol. 2002; 7:275–281.

12. López-Alarcón M, Villalpando S, Fajardo A. Breast-feeding lowers the frequency and duration of acute respiratory infection and diarrhea in infants under six months of age. J Nutr. 1997; 127:436–443.

13. Morrow AL, Rangel JM. Human milk protection against infectious diarrhea: implications for prevention and clinical care. Semin Pediatr Infect Dis. 2004; 15:221–228.

14. Wright AL, Bauer M, Naylor A, Sutcliffe E, Clark L. Increasing breastfeeding rates to reduce infant illness at the community level. Pediatrics. 1998; 101:837–844.

15. Isaacs CE. Human milk inactivates pathogens individually, additively, and synergistically. J Nutr. 2005; 135:1286–1288.

16. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009; 457:480–484.

17. Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009; 9:259.

18. Kim BS, Kim JN, Yoon SH, Chun J, Cerniglia CE. Impact of enrofloxacin on the human intestinal microbiota revealed by comparative molecular analysis. Anaerobe. 2012; 18:310–320.

19. Jeon YS, Chun J, Kim BS. Identification of household bacterial community and analysis of species shared with human microbiome. Curr Microbiol. 2013; 67:557–563.

20. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–721.

21. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011; 27:2194–2200.

22. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009; 75:7537–7541.

23. Rijkers GT, de Vos WM, Brummer RJ, Morelli L, Corthier G, Marteau P. Health benefits and health claims of probiotics: bridging science and marketing. Br J Nutr. 2011; 106:1291–1296.

24. Chang JY, Shin SM, Chun J, Lee JH, Seo JK. Pyrosequencing-based molecular monitoring of the intestinal bacterial colonization in preterm infants. J Pediatr Gastroenterol Nutr. 2011; 53:512–519.

25. Heikkilä MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003; 95:471–478.

26. Fan W, Huo G, Li X, Yang L, Duan C. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. J Microbiol Biotechnol. 2014; 24:133–143.

27. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011; 17:478–482.

28. Tannock GW, Lawley B, Munro K, Gowri Pathmanathan S, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D, Hodgkinson AJ. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol. 2013; 79:3040–3048.

29. Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983; 72:317–321.

30. Lilly DM, Stillwell RH. Probiotics: growth-promoting factors produced by microorganisms. Science. 1965; 147:747–748.

31. Coppa GV, Gabrielli O, Zampini L, Galeazzi T, Ficcadenti A, Padella L, Santoro L, Soldi S, Carlucci A, Bertino E, Morelli L. Oligosaccharides in 4 different milk groups, Bifidobacteria, and Ruminococcus obeum. J Pediatr Gastroenterol Nutr. 2011; 53:80–87.

32. Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012; 18:Suppl 4. 12–15.

33. Conway PL. Selection criteria for probiotic microorganisms. Asia Pac J Clin Nutr. 1996; 5:10–14.
34. Bezirtzoglou E, Romond MB, Romond C. Regulation of the bacterial intestinal implantation in infant born by caesarean section. Comp Immunol Microbiol Infect Dis. 1992; 15:71–74.





 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download