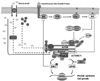
mTOR signaling pathway
Rapamycin, an antifungal macrolide purified from Streptomyces hygroscopicus (Abraham & Wiederrecht, 1996) has potent immunosuppressant and antiproliferative properties and currently is used as a FDA-approved immunosuppressant and anticancer drug. Its cellular target was identified from yeast genetic screening in which mutation of TOR1-1 and TOR2-1 genes showed resistance to the growth-inhibitory properties of rapamycin (Heitman et al., 1991). TOR homologs have also been identified in plants (AtTOR in Arabidopsis thaliana) (Menand et al., 2002), fungus (TOR1 in Cryptcococcus neoformans) (Cruz et al., 1999), Caenorhabditis elegans (CeTOR) (Hara et al., 2002), Drosophila (dTOR) (Oldham et al., 2000; Zhang et al., 2003), and mammals (mTOR) (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994). Unlike yeast TOR1 and TOR2, however, mammals have only one TOR gene.
TOR is a serine/threonine protein kinase and a member of phosphatidylinositol kinase-related kinase (PIKK) family (Schmelzle & Hall, 2000). It consists of 20 tandem HEAT repeats at the N-terminal followed by an FAT and FRB (FKBP12/rapamycin binding) domains. mTOR kinase domain is located in between FRB and FATC (FAT C-terminus) domain at the C-terminus of the protein (Inoki et al., 2005a). Rapamycin binds with immunophilin FKBP12 (FK506-binding protein 12 kDa) in the cell and forms a complex (Abraham & Wiederrecht, 1996). It appears that this FKBP12-rapamycin complex binds to FRB domain and inhibits physiological functions of mTOR, however, exact mechanism has not been elucidated yet. mTOR exists in two distinct protein complexes, mTOR complex1 (mTORC1) and mTOR complex2 (mTORC2) (Hara et al., 2002; Kim et al., 2002; Jacinto et al., 2004; Sarbassov et al., 2004). mTORC1 consists of mTOR, Raptor, mLST8 (GβL), PRAS40 (proline-rich Akt/PKB substrate 40 kDa), and recently identified FKBP38 (Bai et al., 2007; Yang & Guan, 2007). mTORC1 regulates the rate of protein synthesis and cell growth in a rapamycin sensitive way (Fig. 1) (Fingar et al., 2002; Hay & Sonenberg, 2004). While in mTORC2, mTOR interacts with Rictor, mLST8, Protor (protein observed with Rictor) (Pearce et al., 2007), and mSin1 (reviewed in (Yang & Guan, 2007)). Unlike mTORC1, mTORC2 activity is not inhibited by rapamycin at least in a short time period (Jacinto et al., 2004; Sarbassov et al., 2004). Substrates of mTORC2 include Akt and SGK (serum and glucocorticoid-inducible kinase) (Garcia-Martinez & Alessi, 2008). mTORC2 also regulates maturation and stability of conventional PKC (Facchinetti et al., 2008; Ikenoue et al., 2008) and has known to be involved in cytoskeletal organization (Loewith et al., 2002; Jacinto et al., 2004). mTORC2 is activated by growth factors such as insulin but not by nutrients.
Raptor is a scaffold protein which recruits substrates to mTOR and knock-down of Raptor abolishes physiological activity of mTOR (Hara et al., 2002; Kim et al., 2002). mLST8 binds to mTOR kinase domain and activates the kinase activity independent of Raptor (Kim et al., 2003). Recent in vivo studies using mLST8-/- MEFs (mouse embryonic fibroblasts), however, showed that an ability of mTOR to phosphorylate its substrates, S6K and 4EBP1, and to interact with Raptor was not impaired in these cells (Guertin et al., 2006), which suggests that mLST8 may not be an indispensable component of mTORC1 function. PRAS40 acts as a negative regulator of mTORC1 either by binding directly to the mTOR kinase domain and inhibits kinase activity (Vander Haar et al., 2007) or by association with Raptor via a TOR signaling motif (TOS motif) in PRAS40, which can cause substrate competition to Raptor (Oshiro et al., 2007; Sancak et al., 2007; Wang et al., 2007). On the other hand, insulin stimulation phosphorylates Thr246 site of PRAS40 and relieves its inhibitory effect on mTORC1, which suggests that PRAS40 mediates growth factor signals to mTORC1. Bai et al. (2007) identified that FKBP38 also acts as a negative regulator of mTORC1 and overexpression of FKBP38 inhibits S6K1 (T389), S6 (S235/236), and 4EBP1 (T37/46) phosphorylation.
The best-characterized protein substrates of mTORC1 are S6K1 and 4EBP1. mTORC1 recruits and phosphorylates S6K1 and 4EBP1 via interaction of Raptor and TOS motif located at the N-terminus and C-terminus of S6K1 and 4EBP1, respectively (reviewed in (Fingar et al., 2002; Hay & Sonenberg, 2004)). Activated S6K1 phosphorylates 40S ribosomal protein S6, which has been suggested to selectively increase the translation of subset of mRNAs containing a terminal oligopyrimidine (TOP) tract at their 5'-end. Since many of the 5'TOP mRNAs encode ribosomal proteins and other translation factors, activated S6K by mTOR is thought to increase ribosome biogenesis. 4EBP1 is a translational repressor. Its unphosphorylated form binds and inhibits eukaryotic translation initiation factor 4E (eIF4E) which binds to the Cap structure (m7GpppN) at the 5'-end of mRNA transcripts. Phosphorylation of 4EBP1 by mTOR dissociates tight 4EBP1 binding from eIF4E from which eIF4E recruits other components of the translation initiation complex and initiates cap-dependent translation, which increases overall translational capacity of the cell. S6K1 also phosphorylates translation inactivating kinase, eukaryotic elongation factor2 (eEF2) kinase, and inactivates its function (Redpath et al., 1996). mTOR activation, therefore, promotes elongation process as well.
The activity of mTORC1 is finely tuned by upstream positive and negative regulators (Hay & Sonenberg, 2004). Rheb (Ras-homolog enriched in brain) small GTPase is a positive regulator of mTORC1 and is indispensable for mTOR activity. It directly binds to the carboxyterminal lobe of the mTOR catalytic domain and modulates the kinase activity (Long et al., 2005a; Sancak et al., 2007). S6K1 and 4EBP1 can not be phosphorylated if Rheb gene is knocked down (Zhang et al., 2003), whereas, overexpression of Rheb stimulates S6K1 and 4EBP1 phosphorylation (Inoki et al., 2003a; Tee et al., 2003). TSC (tuberous sclerosis complex) 1 and 2 heterodimer acts as Rheb GAP (GTPase activation protein) and converts Rheb to GDP-bound inactive form (Garami et al., 2003; Inoki et al., 2003a; Tee et al., 2003). TSC1/2 integrates a variety of signals including insulin, growth factors, and energy status and negatively regulates mTORC1 (Inoki et al., 2003b). Recently, Drosophila TCTP (translationally controlled tumour protein) has been reported as dRheb GEF (GTPase exchange factor) and activates dTOR (Hsu et al., 2007). Whether mammalian TCTP also can act as Rheb GEF or not needs to be examined.
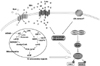
Mechanisms of amino acid sensing
The first indication that amino acids are involved in mTORC1 pathway was shown in the study of hepatocyte autophagy (Blommaart et al., 1995) in which cellular organelles are encompassed by intracellular double membrane followed by formation of autophagosome-lysosome complex and degraded (Shitani & Klionsky, 2004). In the study, addition of amino acids inhibited valine release as a result of autophagic protein degradation in the cell and this inhibition was sharply correlated with the increase of S6 phosphorylation. Moreover, the effects of the amino acids were inhibited by rapamycin. Then the question is how the level of amino acids is sensed and signals to mTORC1 (Fig. 2).
Amino acid sensing on the cell membrane
Amino acids are imported to the cells via various amino acid transporters on the cell membrane. Miotto et al. (1992; 1994) tested the hypothesis whether amino acid signaling is initiated from the membrane transporter using nontransportable peptide Leu8-MAP which contains eight leucine (the single most effective amino acid) residues outside and a leucine analogue, Isovaleryl-L-Carnitine (IVC). Treatment of isolated rat hepatocytes with these compounds inhibited macroautophagy and proteolysis, which indicates that Leu8-MAP, IVC, and probably free leucine must be recognized at the plasma membrane or within an associated vacuole. Different group, however, reported that only after over 1 hour treatment of the Leu8-MAP could phosphorylate S6 in some degree and moreover, in rat adipocytes, S6K1 and 4EBP1 phosphorylation was unresponsive to Leu8-MAP or IVC (Lynch et al., 2000). Another concern on this hypothesis is that amino acid transporter systems have low specificity or strictness in the kind of amino acids or in structural requirement (Hyde et al., 2003). For example, system L amino acid transporter can import isoleucine, valine, methionine, phenylalanine, and histidine to the cells other than leucine. Conversely, pretreatment of 2-amino-3-norbane carboxylic acid (BCH), a selective system L amino acid transporter inhibitor, did not inhibit leucine-induced mTORC1 activity because leucine could be imported through other transporter system (Lynch et al., 2000). On the other hand, it has been suggested that amino acids-induced cell swelling is a mechanism of amino acids-mediated mTORC1 activation. Some amino acids are transported into the cell via Na+-dependent transporters from which intracellular osmolarity is increased due to imported Na+ and accumulated amino acid metabolites (Hyde et al., 2003). Cell swelling itself has shown to exert similar effects as insulin in which protein synthesis is stimulated and protein degradation is inhibited (reviewed in (Haussinger, 1996)). So far, however, how cell swelling regulates these processes is not known. Moreover, the most effective leucine transporter, system L amino acid transporter, is Na+-independent. This suggests that intracellular amino acid concentration or intracellular receptor, but not a particular membrane amino acid transporter, is responsible for the regulation of mTORC1 signaling pathway.
Amino acid sensing by aminoacyl tRNA formation
Bacteria and yeast recognize amino acids starvation by increased concentration of deacylated tRNA or reduced availability of aminoacylated tRNA. In Jurkat cells, amino acid alcohols which compete with amino acids prevent aminoacyl-tRNA formation and inhibit S6K1 phosphorylation (Iiboshi et al., 1999). Inhibition of protein synthesis by applying translation inhibitors such as cycloheximide (CHX) and puromycin has shown to increase S6K1 phosphorylation in the amino acids starvation condition (Iiboshi et al., 1999). These data suggest that increased level of intracellular amino acids resulted from translation inhibition and of aminoacylated tRNA is responsible for the amino acid sensing in mammalian cells as well. However, this is not consistent with time frame in which mTORC1 is activated. The level of intracellular amino acids is not changed during short time (30 min) amino acids starvation or stimulation, whereas, S6K1 phosphorylation is changed within this short time frame. Moreover, in rat adipocytes, leucine alcohol (leucinol) treatment did not affect 4EBP1 phosphorylation (Lynch et al., 2000). On the other hand, it appears that Rheb and TOR are not involved in nutrient import. Suppression of Rheb and TOR gene expression by RNAi in Drosophila S2 cells did not affect nutrients (glucose & bulk amino acids) import but significantly reduced ribosome biogenesis, protein synthesis, and cell size (Hall et al., 2007). Taken together, these data raise a question on this hypothetical mechanism as well.
Amino acid sensing by amino acid metabolites
Imported amino acids are subjected for amino acid metabolism and there are several reports suggesting that amino acid metabolites including ATP are responsible for mTORC1 activation. For example, leucine is metabolized to α-ketoisocaproic acid (α-KIC) by reversible aminotransferase (AT), which is then further metabolized to TCA cycle intermediates by branched-chain α-ketoacid dehydrogenase (BCKDH). Otherwise, leucine as it is can be an allosteric activator of glutamate metabolizing enzyme, glutamate dehydrogenase (GDH) (Fahien et al., 1988; Sener & Malaisse, 1980). The consequence of leucine metabolic pathways would be production of ATP and other metabolically-linked secondary signals, which was proposed as a mechanism of mTOR signaling pathway activation by leucine in pancreatic β-cells (McDaniel et al., 2002). In fact, Dennis et al. (2001) have previously reported that mTOR is an intracellular ATP sensor and ATP depletion caused by incubation of cells with 2-deoxyglucose or several mitochondria inhibitors including rotenone and antimycin A inhibits S6K1 and 4EBP1 phosphorylation (Inoki et al., 2003b; Xu et al., 2001). A concern on this hypothesis is that the concentration of ATP is not affected by short time amino acids deprivation or stimulation in most mammalian cells (Dennis et al., 2001). Nevertheless, it has been shown that an AT inhibitor, aminooxyacetic acid (AOAA), inhibits S6K1 phosphorylation upon leucine treatment in pancreatic β-cells (Xu et al., 2001). A nonmetabolized leucine analog, β (±) 2-aminobicyclo [2.2.1] heptane-2-carboxylic acid, also failed to stimulate S6K1 phosphorylation in the absence of leucine (McDaniel et al., 2002). Moreover, proteolytic dose responses and inhibition of autophagy to L-leucine and to IVC, readily metabolizable leucine analogue, were similar in the perfused rat liver (Miotto et al., 1992). Stimulation with α-KIC phosphorylates 4EBP1 in the absence of leucine in adipocytes (Lynch et al., 2000). Taken together, this suggests that although it is difficult to pinpoint exactly what it is, certain products of leucine oxidation are responsible for mTORC1 activation.
In the liver, however, leucine is poorly metabolized owing to its low rate of transamination. Therefore, we still can not exclude a possibility that the effect of leucine on the mTORC1 activation is due to leucine itself at some undefined locus of recognition. Leucine metabolizing enzyme, AT, is a reversible aminotransferase which also can convert α-KIC to leucine. Interestingly, α-KIC induced phosphorylation of 4EBP1 was less than leucine induced 4EBP1 phosphorylation and moreover, α-KIC effect was attenuated in the presence of transaminase inhibitor, L-cycloserine (Lynch et al., 2000). Lynch et al. (2000) also found that norleucine which is not incorporated in protein translation was as effective as leucine in 4EBP1 phosphorylation, which suggests that there may be a specific intracellular leucine receptor, which recognizes the specific structure of leucine. However, leucine analogues which contain either strong leucine agonist or antagonist activity in one cell line do not always exert the same effect in different cell lines. Therefore, this proposed mechanism can be cell-type specific.
Amino acid sensing by mTORC1
Although the mechanism is not known, it is obvious that amino acids regulate mTORC1 activity (Hara et al., 1998). That is, amino acids stimulation increases S6K1 and 4EBP1 phosphorylation, whereas, amino acids deprivation rapidly dephosphorylates S6K1 and 4EBP1. Therefore, extensive research efforts have centered on mTORC1 regulation itself.
Ectopic overexpression of Rheb completely inhibited S6K1 and 4EBP1 dephosphorylation upon amino acids withdrawal (Long et al., 2005a; 2005b; Saucedo et al., 2003). Since Rheb activates mTORC1 activity by directly binding to mTOR, Rheb-mTOR interaction was examined. Long et al. (2005b) showed that Rheb-mTOR interaction was in fact reduced upon amino acids withdrawal, whereas the interaction was fully recovered when amino acids alone were added to D-PBS. Amino acids, however, did not change Rheb-Raptor and Rheb-mLST8 interaction (Long et al., 2005b). Curiously, however, Rheb-mTOR interaction is not affected by nucleotide binding status of Rheb (Long et al., 2005a). The binding of nucleotide-free mutant of Rheb to mTOR was even stronger than that of wild-type Rheb, although it impaired mTOR kinase activity. Amino acids modulation of nucleotide binding status of Rheb is also not in consistency. For example, GTP level of overexpressing Rheb was increased by amino acids stimulation (Smith et al., 2005), but endogenous Rheb-GTP charging was not changed by amino acids withdrawal (Nobukuni et al., 2005). Roccio et al. (2006) reported that amino acids starvation did not change Rheb-GTP level, but add back amino acids clearly increased Rheb-GTP loading. In consistent with this, TSC1/2 is also not required for amino acids-mediated mTORC1 regulation (Roccio et al., 2006; Smith et al., 2005). Amino acids withdrawal still dephosphorylated S6K1 and 4EBP1 independent of TSC2 gene expression (Nobukuni et al., 2005; Smith et al., 2005). On the other hand, it has been proposed that amino acids regulate mTORC1 activity by modulating mTOR-Raptor interaction. Amino acids starvation causes tighter binding of Raptor to mTOR which inhibits mTOR kinase activity whereas amino acids stimulation loosens the interaction (Kim et al., 2002). The mLST8-mTOR interaction was not affected by amino acids. This suggests that amino acids specifically modulate mTOR-Raptor interaction and regulate mTOR kinase activity.
Amino acid sensing by intracellular proteins
Recently, several intracellular proteins have been identified as intracellular amino acid sensors which include hVps34, MAP4K3, and Rag.
Vps34: Vps34 is a Class III PI3K which forms an active complex with another kinase Vps15 and generates PtdIns(3)P by phosphorylating the 3' hydroxyl position of the phosphatidylinositol ring (reviewed in (Backer, 2008; Nobukuni et al., 2007; Yan & Backer, 2007)). Vps34 was initially known to play an important role in vesicular trafficking (Lindmo & Stenmark, 2006; Odorizzi et al., 2000) and a role of Vps34 in amino acids sensing has been recently reported (Byfield et al., 2005; Nobukuni et al., 2005). That is, hVps34 activity as well as PtdIns(3)P production was increased by amino acids stimulation. On the contrary, suppression of hVps34 gene expression with hVps34 siRNA or overexpression of GFP-FYVE (Fab1p, YOTB, Vac1p, EEA1 (early endosomal antigen 1)) expression vector which acts as a dominant negative by sequestering PtdIns(3)P impaired S6K1 and 4EBP1 phosphorylation but did not affect insulin-stimulated Akt phosphorylation. It appears that, however, amino acids do not directly regulate hVps34 activity. Instead, amino acids stimulation facilitates Ca2+ influx, which in turn increases the interaction of Ca2+/CaM with the hVps34-mTOR and activates mTORC1 (Gulati et al., 2008). There are several concerns, however, on presenting hVps34 as an amino acids sensing system to activate mTORC1. Ectopic expression of hVps34 to mammalian cells increased S6K phosphorylation only in the presence, but not in the absence of amino acids (Nobukuni et al., 2005). In the study of Yan et al. (2009) hVps34 activity was not affected by Ca2+ chelators or CaM inhibitors. Most of all, in a recent Drosophila in vivo study, loss-of-function mutant animals of Drosophila Vps34 showed disrupted autophagy and endocytosis but TOR signaling was not affected by disruption of Vps34 gene (Juhasz et al., 2008). That is, cell size was similar between Vps34 mutant and wild-type cells and moreover, modulation of cell size by overexpressing Rheb (increased cell size) or TSC1/2 (decreased cell size) was not affected by Vps34 mutation. Nevertheless, amino acids starvation induced autophagy in control animals but not in Vps34 mutant animals. Considering that reported function of hVps34 in amino acid signaling has been all performed in cell culture system, this observation in vivo raises a significant question on the proposed model.
MAP4K3: Findlay et al. (2007) identified MAP4K3 protein kinase as an important intracellular protein in amino acids sensing. They showed MAP4K3 kinase activity was regulated by amino acids and overexpression of MAP4K3 specifically increases S6K phosphorylation even in the amino acids starvation condition in a TSC1/2 independent manner. This is a difference between hVps34 and MAP4K3 in amino acids sensing since ectopic expression of hVps34 increases S6K1 phosphorylation only in the presence of amino acids. Nevertheless, it would be interesting to address a question whether overexpression of MAP4K3 increases PtdIns(3)P either in the presence or absence of amino acids.
Rag: Most recently, two independent research groups newly discovered that Rag (Ras-related GTPases) small GTPases (RagA, B, C, and D) play an important role in amino acid signaling (Kim et al., 2008; Kim & Guan, 2009; Sancak et al., 2008).
In Drosophila S2 cells, knock-down of dRag gene expression impaired dS6K phosphorylation upon amino acids stimulation (Kim et al., 2008). In mammalian cells, expression of GTP-bound active RagAQL/RagBQL overcomes S6K1 dephosphorylation in amino acids deprivation condition, whereas GDP-bound inactive RagATN/BTN inhibited S6K1 phosphorylation upon amino acids stimulation. Moreover, dRagAQL expressing cells showed bigger cell size than neighboring wild type cells and inhibited starvation-induced autophagy in Drosophila. Sancak et al. (2008) identified RagC as a Raptor interacting protein using a purification method. Amino acids increase GTP-level of RagB and RagBQL-RagCSN heterodimer increases mTORC1 activity by increasing Raptor-mTOR interaction. On the other hand, in RagBQL expressing cells, mTOR was already present on the Rab7 positive peri-nuclear and vesicular structures even in the amino acids deprivation condition. Interestingly, hVps34 and hVps15 also co-localize to Rab7-positive late endosome. This suggests that the function of Rag, mTOR, and hVps34 can be interconnected on Rab7 positive endosome membrane and may involve in amino acids transporter trafficking to the cell membrane as yeast Rag homologue, Gtr does (Gao et al., 2006).
Recently, Nicklin et al. (2009) found that L-glutamine uptake by solute carrier family1 (SLC1A5) transporter is important for L-leucine/essential amino acids (EAA) import into the cells. Simultaneous efflux of L-glutamine out of cells through SLC7A5/SLC3A2, a bidirectional transporter, facilitates L-leucine/EAA import into the cells from which mTORC1 is activated. In contrast to L-glutamine, L-glutamic acid and α-ketoglutarate pretreatment could not activate S6K1 upon EAA treatment, which suggests that L-glutamine effect on S6K1 is not due to increasing cellular energy level. Interestingly, previously performed large-scale genetic array analysis identified that protein mutations involved in glutamate/glutamine homeostasis also genetically interact with Gtr2 (Dubouloz et al., 2005). Lethal phenotype of these mutants was reversed either by glutamate supplement in the medium or by introduction of Gtr2-expression plasmids. Moreover, Gtr2 mutant suppressors identified from genome-wide screen include genes which are directly or indirectly involved in glutamate/glutamine synthesis/degradation. It has been shown that Gse complex consisted of five proteins encoded by Gtr1, Gtr2, YBR077C (EGO3), YKR007W (EGO1), and LTV1 genes plays an important role in recycling of general amino acid permease, Gap1p, from the late endosome to the plasma membrane in yeast (Gao et al., 2006). Therefore, it will be interesting to address a question whether Rag affect SLC1A5 and SLC7A5/SLC3A2 transporter trafficking and whether imported L-leucine/EAA activates mTORC1 through Rag.
Concluding remarks
How amino acids regulate protein synthesis and cell growth has been a fundamental biological question in the field. Since inhibition of TOR pathway mimics amino acids starvation condition and moreover, amino acids are the most potent activator of TOR, extensive research efforts have centered on elucidating a mechanism of amino acids sensing in mTOR signaling pathway. From the cell membrane to intracellular proteins, each path of amino acids has been examined for a possible amino acid sensing point, however, there have been still many controversies in experimental data due to such a structural and metabolic diversity of amino acids. Recent identification of hVps34, MAP4K3, and Rag GTPases as important intracellular proteins in amino acid signaling and of a mechanism of L-Leucine/EAA import into the cells made a significant progress in our understanding of amino acids-mediated mTOR activation. However, how imported amino acids are sensed by hVps34, MAP4K3, or Rag GTPases and activate mTORC1 still remains as an open question. The importance of understanding amino acid signaling is that it has many significant physiological and pathological implications in vivo. Anabolic response of the body after protein meal ingestion is closely correlated with phosphorylation of S6K1 and 4EBP1 in mice, rat, and human (Balage et al., 2001; Long et al., 2000; Shah et al., 2000). Insulin production in pancreatic β-cells is increased by amino acids stimulation (May & Buse, 1989) and amino acids infusion increases the sensitivity of muscle protein synthesis to insulin (Garlick & Grant, 1988). Furthermore, activated S6K in turn phosphorylates and degrades insulin receptor substrate (IRS) and inhibits PI3K signaling pathway as a negative feedback inhibition, which can decrease insulin sensitivity and lead to diabetes (Fig. 1). Overactivated mTORC1 signaling pathway is frequently found in various cancers and tissue/organ hypertrophy as well (Guertin & Sabatini, 2007; Inoki et al., 2005b; Lee et al., 2007). Although rapamycin has been used as an FDA-approved drug to treat some of the cancers including sarcoma, suppression of S6K phosphorylation can result in activating PI3K signaling which can further exacerbate the disease state. Therefore, if we elucidate a mechanism on the amino acid signaling pathway to mTORC1 activation, more specific and effective way to prevent or treat these diseases can be developed.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download