Abstract
BACKGROUND/OBJECTIVES
Nelumbo leaves have been used in traditional medicine to treat bleeding, gastritis, hemorrhoids, and halitosis. However, their mechanisms have not been elucidated.
MATERIALS/METHODS
The present study prepared two Nelumbo leaf extracts (NLEs) using water or 50% ethanol. Inflammatory response was induced with LPS treatment, and expression of pro-inflammatory mediators (inducible nitric oxide synthase, cyclooxygenase-2, tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6 and nitric oxide (NO) and prostaglandin E2 (PGE2) productions were assessed. To determine the anti-inflammatory mechanism of NLEs, we measured nuclear factor-κB (NF-κB) activity. Major metabolites of NLEs were also analyzed and quantified.
Inflammation is a natural response that promotes the survival of a host in the presence of a variety of internal and external insults [1]. However, abnormal regulation of inflammatory processes can result in the destruction of cells, or may disturb cellular metabolism and thus contribute to the development of chronic diseases. Macrophages play a central role in the innate immune response and chronic inflammation processes by secreting pro-inflammatory cytokines such as interleukin (IL)-6 and tumor necrosis factor-α (TNF-α) [12]. Macrophages activate inflammation-related genes that are regulated via nuclear factor-κB (NF-κB) signaling [3]. Activated NF-κB translocates to the nucleus, where it induces the expression of pro-inflammatory mediators and cytokines, including inducible nitric oxide synthase (iNOS), cyclooxygenase-2 (COX-2), TNF-α, IL-1β, and IL-6 [34]. Expression of these proteins elicits further immune responses. Recent studies have identified sustained activation of signaling pathways relevant to inflammation in various pathological conditions, suggesting that this is a crucial underlying cause of many chronic diseases such as atherosclerosis, cardiovascular disease, autoimmune disease, rheumatoid arthritis, cancer, and diabetes [56].
The immunomodulatory effects of plant extracts have been explored to investigate whether these could prevent chronic diseases that are caused by overproduction of pro-inflammatory mediators. The Nelumbo nucifera Gaertn belongs to the Nymphaeaceae family and is an aquatic perennial plant that is widely cultivated throughout Eastern Asia, Australia, and India for food, beverages, ornamental, and therapeutic purposes [7]. All parts of the Nelumbo, including the leaves, fruits, flowers, rhizomes, and seeds, have been reported to have pharmacological effects [89]. Nelumbo leaves have been used as a vegetable and a functional food, as well as to make tea and alcoholic beverages in Korea [910]. The Nelumbo leaf is used in folk medicine to treat bleeding, gastritis, hemorrhoids, mouth inflammation, and halitosis [111213]. The health-promoting effects of Nelumbo in diabetes, cancer, hepatoxicity, and obesity have been associated with its antioxidant activity [1415161718]. This has been attributed to the presence of flavonoids and other phenolic substances. Nelumbo leaves contain quercetin, myricetin, kaempferol, diosmetin, and isorhamnetin derivatives [819]. The catechol structure within flavonoids is a hydrogen atomdonating group that scavenges reactive oxygen species (ROS) and thereby exerts antioxidant effects [20]. For instance, Nelumbo leaf extract (NLE)-derived quercetin was demonstrated to be a potent flavonoid that prevented low-density lipoprotein oxidation [14]. It was also suggested to exert antibacterial activity [12]. However, even though Nelumbo leaf is commonly administered as a tea or alcoholic beverage, its active compounds have generally been analyzed using methanol or ethyl acetate extracts. Since the solvent properties determine which compounds are extracted, the active compounds administered in aqueous extracts may differ from those analyzed in organic solvent extracts.
Metabolomics is an emerging tool for non-targeted profiling and identification of all metabolites in a sample and it is being applied to various comprehensive plant analyses [21]. The resulting data are subjected to multivariate statistical analysis using principal component analysis (PCA), partial least-squares discriminatory analysis (PLS-DA), or orthogonal PLS-DA (OPLS-DA) to identify different groups and screen for effective discriminants. This technique can provide a useful tool for the identification and comparison of plant extract constituents.
In this study, we investigated and compared the anti-inflammatory effects of water- or ethanol-based NLEs. In addition, we determined the possible active compounds present in these extracts.
Nelumbo leaves were purchased from Dadobang (Seoul, South Korea). Steamed and dried leaves were extracted with either 50% ethanol or water. These extracts were filtered, rotary evaporated, and lyophilized. The resulting Nelumbo leaf ethanol extract (NLEE) or water extract (NLWE) was dissolved in dimethyl sulfoxide or media and stored at −20℃ until use.
Murine macrophage RAW 264.7 cells were purchased from the Korea Cell Line Bank (Seoul, Korea). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% inactivated fetal bovine serum (v/v), 5% penicillin (100 U/ml), and streptomycin (100 µg/ml; Invitrogen, Carlsbad, CA, USA) and maintained at 37℃ in a humidified atmosphere of 5% CO2. Either NLEE or NLWE (1, 10, or 50 µg/ml) was incubated with the plated cells for 24 h prior to the addition of 1 µg/ml of lipopolysaccharide (LPS; Sigma-Aldrich, St. Louis, MO, USA) for the indicated time-period.
Cells were pre-incubated with the indicated concentrations of either NLEE or NLWE for 24 h and then stimulated with 1 µg/ml LPS for 18 h. Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich, St. Louis, MO, USA). Briefly, cells were incubated with MTT solution for 2 h at 37℃ prior to removing the medium and lysing the cells using isopropanol. The absorbance of the colored solution at 450 nm was quantified by a Spectra MAX 340 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
RAW 264.7 cells were pre-treated with either NLEE or NLWE (1, 10, or 50 µg/ml) for 24 h and then induced with LPS (1 µg/ml) for 18 h. The supernatants was collected and the levels of nitrite and PGE2 were determined by a nitrate/nitrite Colorimetric Assay kit and a PGE2 enzyme immunoassay kit (Cayman Chemical, Ann Arbor, MI, USA), in accordance with the manufacturer's protocol.
Cells were pre-treated with either NLEE or NLWE and then stimulated with LPS for 18 h prior to collecting the cell cultures media and analyzing the levels of TNF-α, IL-1β, and IL-6 using enzyme-linked immunosorbent assay kits (R&D systems, Minneapolis, MN, USA).
RAW 264.7 cells were incubated with either NLEE or NLWE for 24 h and then induced by LPS (1 µg/ml) for 4 h. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions and stored at -80℃ until use. Total RNA was reverse-transcribed using a Superscript First Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). All PCR analyses were subsequently carried out using a GenePro Thermal Cycler (Bioer Technology Co. Ltd., Hangzhou, China). After an initial denaturation at 95℃ for 5 min, 25-30 amplification cycles were conducted in the following order: denaturation at 95℃ for 30 sec, annealing at varying temperatures for 30 sec, and elongation at 72℃ for 1 min. A final extension step was carried out at 72℃ for 10 min. After amplification, PCR products were separated by electrophoresis on a 1% agarose gel and visualized under UV light.
After NLEE or NLWE pre-treatment, cells were stimulated with LPS for either 15 min or 18 h and then lysed in RIPA lysis buffer containing cocktail protease inhibitors and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA). Cell debris was removed by centrifugation for 30 min at 10,000 g at 4℃. Protein concentrations were determined using a Bradford Assay kit (Bio-Rad, Hercules, CA, USA), according to the manufacturer's instructions. Lysates were boiled in sample buffer for 5 min. Equivalent amounts of sample protein were then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20 for 1 h at room temperature, and then probed with antibodies against iNOS, COX-2, phosphorylated inhibitor of NF-κB (Iκ-B)-α, and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The membranes were washed and then exposed to horseradish peroxidase-conjugated secondary antibodies (1:4000 dilutions). Immunoreactive bands were detected by enhanced chemiluminescence solution (Animal Genetics, Seoul, South Korea) and exposed to X-ray film.
Cells were harvested after a 24-h pre-treatment with NLEE or NLWE and 20-min LPS stimulation. Nuclear proteins were extracted using a Nuclear Extraction kit (Cayman, Ann Arbor, MI, USA), and NF-κB transcriptional activity was measured using an NF-κB Transcription Factor Assay kit, according to manufacturer's instructions (Cayman, Ann Arbor, MI, USA).
NLEs were analyzed using a high-performance liquid chromatography (HLPC) system with a PDA Detector (Accela 80 Hz) and an LTQ-Velos ion trap mass spectrometer fitted with a heat electrospray ionization interface (Thermo Fisher Scientific, Waltham, MA, USA) with a C18 column (1.9 µm, 2.0 × 100 mm, UPLC BEH C18, Waters, USA). The elution gradient was carried out with a binary solvent system consisting of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) at a constant flow rate of 0.3 ml/min. The linear gradient profile was as follows: 5-10% solvent B over 5 min, 10% solvent B for 10 min, 10-40% solvent B over 30 min, 40-90% solvent B over 40 min, then returned to 5% solvent B over 5 min, followed by conditioning for 10 min, finished at 55 min. Acquisition was performed in the negative and positive ion modes using electrospray ionization. The capillary temperature was 275℃, with sheath gas at 35 units and auxiliary gas at 5 units. NLEs were run in triplicate and the collected data was processed for profiling and identification.
Peak detection, alignment, and identification were performed using the SIEVE software (Thermo Fisher Scientific, Waltham, MA, USA). The acquired results were analyzed by multivariate statistical analysis using SIMCA (Umetrics, Umea, Sweden). PCA was used to visualize the similarities and differences between NLEE and NLWE, and OPLS-DA was applied to detect maximal separation of distinctive metabolites in each partially purified fraction. The MS/MS fragmentation patterns, retention time, and UV-visible spectra were used for informative non-targeted metabolic profiling and the acquired LC-MS/MS spectrum was compared with those in the MassBank database (www.massbank.jp), KEGG database (www.kegg.jp), Metlin (http://masspec.scripps.edu), and other published reports.
Quantification of key metabolites was conducted using a standard curve generated from authentic standards of quercetin and catechin. Analysis was performed by HPLC (Dionex HPLC-3000, Dionex Corp., Sunnyvale, CA, USA) with UV detector (Ultimate 3000 Diode array detector, Dionex Corp., Sunnyvale, CA, USA) using X-bridge C18 column (3.5 um, 4.6 × 150 mm. Waters) at 35℃. The solvents were 0.1% formic acid in water (phase A) and acetonitrile (phase B) at flow rate of 1 mL/ml. The linear gradient profile was as follows: 5-10% B in 5 min, held at 10% B in 5-10 min, 10-40% B in 10-30 min, finished at 30 min. UV spectrum were measured at 275 nm.
All data presented were expressed as the mean ± standard error (SE) of three or more independent experiments. Statistical comparisons were made using analysis of variance (ANOVA), followed by Duncan's multiple range test. In all cases, results were considered significant if the P-value was less than 0.05. All statistical tests were performed using SPSS statistical analysis software (Chicago, IL, USA).
RAW 264.7 cells were treated with various concentrations of NLEE or NLWE for 24 h prior to examining their viability using MTT assays. There were no significant changes in cell viability (Fig. 1), indicating that neither NLEE nor NLWE was cytotoxic at concentrations of up to 50 µg/ml.
We investigated whether NLEs could modulate NO production in activated RAW 264.7 cells. The cells were pre-treated with NLEE or NLWE (1, 10, or 50 µg/ml) and stimulated with 1 µg/ml of LPS for 18 h prior to determination of the levels of nitrite and iNOS expression. LPS stimulation significantly increased nitrite production by about 3-fold, as compared to unstimulated control cells (Fig. 2A). However, 50 µg/ml of NLEE (IC50 of 201.4 µg/ml) and NLWE (IC50 of 231.2 µg/ml) significantly suppressed LPS-stimulated nitrite production, and iNOS mRNA and protein expressions (Fig. 2B and C). NLWE, at a concentration of greater than 10 µg/ml, significantly reduced iNOS mRNA levels in a concentration-dependent manner (Fig. 2B). In addition, NLEE and NLWE significantly suppressed iNOS protein expression, as compared to the LPS-stimulated control cells. These results indicated that NLEE and NLWE reduced nitrite production and iNOS expression in a concentration-dependent manner.
To test the effects of NLEs on pro-inflammatory mediators, PGE2 and COX-2 levels were assessed in RAW 264.7 cells incubated with NLEE or NLWE for 24 h prior to LPS stimulation for either 4 h or 16 h. NLEE (IC50 of 5.3 µg/ml) significantly reduced PGE2 production at a concentration of greater than 1 µg/ml (Fig. 3A). NLWE (IC50 of 4.8 µg/ml) also significantly decreased LPS-induced PGE2 production in a concentration-dependent manner (Fig. 3A). The lowest concentration of NLWE that reduced PGE2 production was 10 µg/ml. Furthermore, NLEE reduced COX-2 mRNA and protein expression. However, NLWE significantly reduced COX-2 protein levels but not its mRNA expression. These results suggested that NLEE reduced the production of the pro-inflammatory mediator, PGE2, and the expression of COX-2 more effectively than NLWE.
We investigated whether NLEs influenced the production of the pro-inflammatory cytokines, IL-1β, IL-6, and TNF-α, as well as their gene expression. NLEE (10 µg/ml) was sufficient to suppress IL-1β levels in RAW 264.7 macrophage cells. IL-1β levels were reduced by about 60% in cells exposed to 50 µg/ml NLEE (IC50 of 38.8 µg/ml), as compared to only 25% reduction in those exposed to 50 µg/ml NLWE (IC50 of 88.8 µg/ml) (Fig. 4A). However, both NLEE and NLWE slightly reduced IL-1β mRNA levels. NLWE, but not NLEE, treatment significantly reduced the levels of IL-6, and its mRNA, as compared to LPS-stimulated control cells (Fig. 4B). Both NLEE and NLWE noticeably inhibited the expression and production of TNF-α (Fig. 4C). NLEE (10 µg/ml) significantly decreased TNF-α mRNA and the secreted forms of TNF-α in a concentration-dependent manner. In addition, 1 µg/ml NLWE (IC50 of 38.1 µg/ml) was enough to reduce the secretion of TNF-α (Fig. 4C). Taken together, these NLEs markedly reduced the LPS-stimulated mRNA expression of TNF-α, IL-1β, and IL-6 genes, as well as these cytokine levels, in a concentration-dependent manner.
Since the NF-κB pathway regulates the expression of inflammatory mediators and pro-inflammatory cytokines, we next investigated whether NLEs modulated NF-κB activity. As shown in Fig. 5A, NLEs reduced NF-κB activity in a concentration-dependent manner. NLEE and NLWE significantly reduced NF-κB activity at concentration greater than 10 and 1 µg/ml, respectively. Furthermore, the level of phosphorylated cytosolic IκB was lower in cells pre-treated with NLE for 24 h (Fig. 5B). NLEE and NLWE reduced phosphorylation of IκB at concentrations greater than 50 and 10 µg/ml, respectively. These results indicated that NLE decreased the transcriptional activity of NF-κB by reducing phosphorylation of IκB proteins, and that NLWE produced a more effective reduction in NF-κB activity than NLEE.
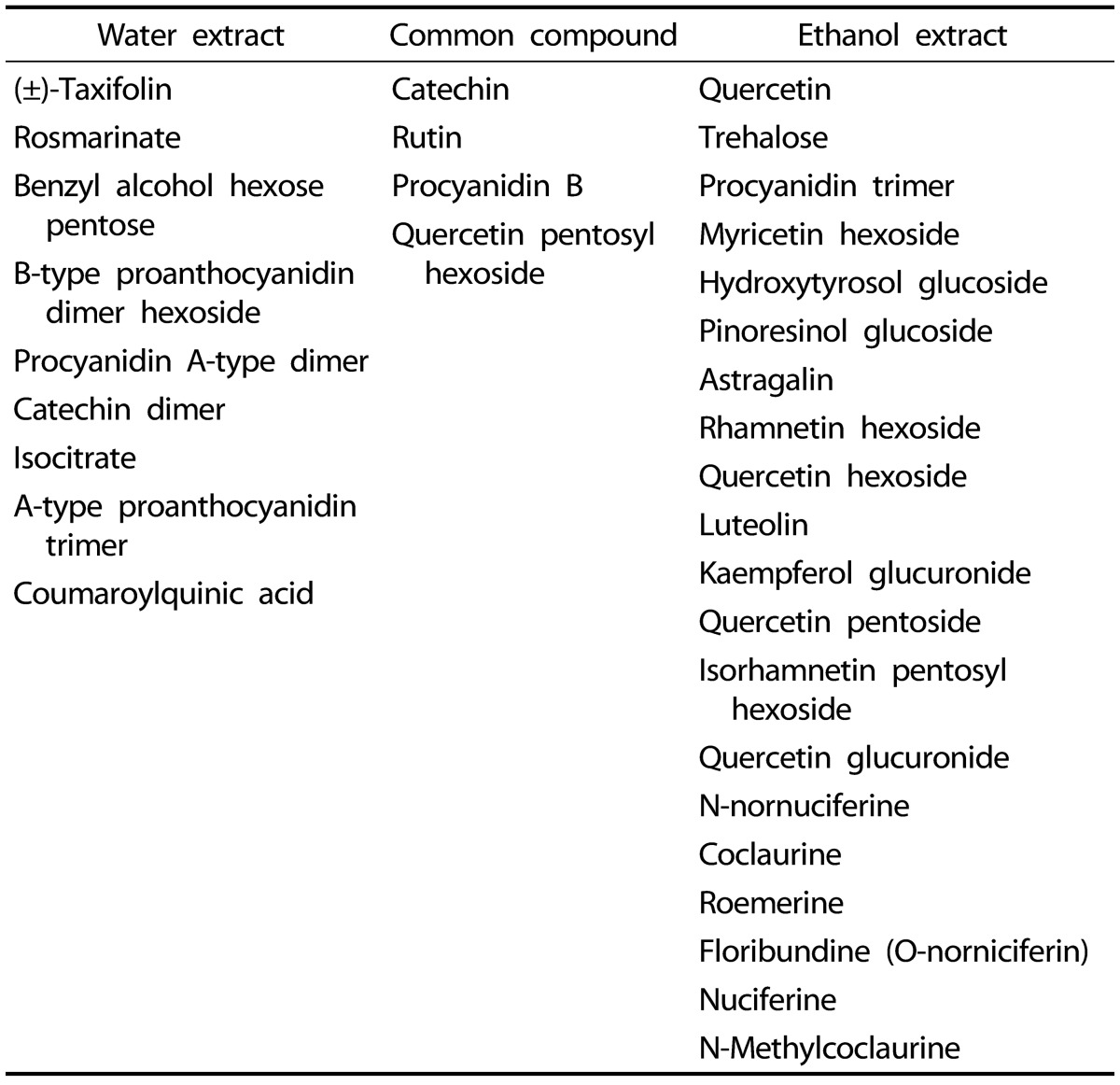
Since NLEs reduced pro-inflammatory mediator and cytokine productions, we determined the major bioactive compounds in NLEs. Multivariate statistical analysis was used to distinguish the metabolites present in NLWE and NLEE. Unsupervised PCA showed a clear differentiation of the metabolites extracted using water and ethanol, as indicated in Fig. 6. The replicates of each extract clustered closely together and were clearly separated into different groups in both of ion detection modes (positive and negative). In negative ion mode, the NLEE metabolites were mainly discriminated from those in the NLWE by component 1 (88.3%), followed by component 2 (10.2%); a higher dependency on component 1 (91.5%) was observed in positive ion mode. Furthermore, the OPLS-DA was used to screen the highly significant metabolites in each extract (Fig. 7). Among 396 metabolites detected by LC-MS/MS analysis in negative ion mode, 57 distinctive metabolites were identified in the NLWE, while 136 were found in the NLEE (P < 0.01). In positive ion mode, these values were 23 in the NLWE and 97 in the NLEE (P < 0.01) (supplementary data). The main compounds identified in each extract are listed and summarized on Table 1. Catechin, rutin, procyanidin B, and quercetin pentosyl hexoside were found in both extracts; these could contribute to the observed anti-inflammatory activities. Since both extracts reduced inflammatory responses, we investigated bioactive compounds which existed in both water and ethanol extracts. The bioactive compounds in both NLE were catechin and quercetin. NLWE and NLEE contained 22.3 and 34.3 µg/mg of catechin and 3.72 and 1.80 µg/mg of quercetin (Fig. 8). Different levels of catechin and quercetin in both extracts could cause anti-inflammatory responses in Raw 264.7 cells.
In this study, we provide evidence that Nelumbo leaf reduced pro-inflammatory mediators and cytokines expression through suppression of NF-κB activity so Nelumbo leaf can be used as an anti-inflammatory agent or functional food to modulate inflammation.
Since NLEs have shown several health benefits, researchers elucidated their major constituents and identified high levels of phenolic compounds such as catechin, quercetin, and proanthocyanidins [2223]. A range of amounts and types of total phenolic compounds were identified, depending on the extraction solvent employed [24]. Since Nelumbo leaf has been used as a tea or alcoholic beverage in Korea, water and 50% ethanol were used as the extraction solvents in the present study and two major constituents, catechin and quercetin, were found. In this study, we showed that NLEE contained more catechin compared to NLWE (34.3 µg/mg vs. 22.3 µg/mg). Since solubility of catechin has been increased by temperature and ethanol concentration [25], NLEE contained more catechin compared to NLWE. Catechin has been shown to have antitumorigenic and anti-atherosclerotic effects [262728] and these health benefits reflect anti-inflammatory effects [293031]. Catechin reduced gastric inflammation by suppressing IL-8 production in gastric cancer and human umbilical vein endothelial cells [29] and decreased COX-2, iNOS, IL-1β, and TNF-α expression by suppressing NF-κB in skin carcinoma cells [31].
One of major constituents in NLEs was quercetin and NLWE contained more than twice of quercetin compared to NLEE. Quercetin also has been shown to have anti-tumor, antioxidant, and anti-inflammatory properties [323334]. Other study showed that quercetin reduced iNOS and COX-2 expression via inhibition of NF-κB signaling pathways [33]. These findings indicate that two major NLE components, catechin and quercetin, could attenuate LPS-induced inflammatory responses.
Although NLWE and NLEE had strong anti-inflammatory effects, NLWE and NLEE showed different effects on pro-inflammatory mediator and cytokines production and expression. NLWE reduced more of iNOS, IL-6, and TNF-α production and NF-κB activity compared to NLEE and NLEE suppressed PGE2 and IL-1β production. These difference could be caused by metabolites NLEs contained. NLWE contains taxifolin, which suppresses inflammatory responses [353637]. Extracts of Ulmus macrocarparoot or Pinus brutia bark, which mainly contained catechin and taxifolin, inhibited NO production by suppressing iNOS and COX-2 expression [3637]. Moreover, taxifolin glycoside significantly reduced atopic dermatitis by effects on iNOS and COX-2 [35] and also ameliorated cerebral ischemia-reperfusion injury by inhibiting NF-κB activity [38]. However, NLEE contains luteolin which suppressed inflammation in several model through JAK/STAT and/or NF-κB pathway [394041]. Luteolin reduced iNOS expression through inhibition of NF-κB translocation in LPSstimulated RAW 264.7 cells [39]. In addition, luteolin inhibited IL-8 production, and COX-2 and iNOS expression by decrease of JAK/STAT pathway [41]. Therefore, different effects of NLEs on pro-inflammatory mediator or cytokine production and/or expression were caused by metabolites they contained.
The present study provided evidence that NLEs reduced LPS-activated macrophage responses via inhibition of NF-κB activation and two major constituents of these extracts were catechin and quercetin. Moreover, the concentration of NLEs required to exert these anti-inflammatory effects did not cause cytotoxicity. Further in vivo studies are warranted to confirm the anti-inflammatory effects of NLEs in people who are susceptible to inflammation-related diseases. This study indicated that NLEs could be used in a therapeutic approach to the prevention or inhibition of diseases characterized by chronic inflammation related to NF-κB activity.
References
1. Hunter M, Wang Y, Eubank T, Baran C, Nana-Sinkam P, Marsh C. Survival of monocytes and macrophages and their role in health and disease. Front Biosci (Landmark Ed). 2009; 14:4079–4102. PMID: 19273336.

2. Laroux FS. Mechanisms of inflammation: the good, the bad and the ugly. Front Biosci. 2004; 9:3156–3162. PMID: 15353345.

3. Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996; 87:13–20. PMID: 8858144.
4. Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005; 4:281–286. PMID: 16101534.

5. Aggarwal BB, Gehlot P. Inflammation and cancer: how friendly is the relationship for cancer patients? Curr Opin Pharmacol. 2009; 9:351–369. PMID: 19665429.

6. McGeer PL, McGeer EG. Inflammation and the degenerative diseases of aging. Ann N Y Acad Sci. 2004; 1035:104–116. PMID: 15681803.

7. Liao CH, Lin JY. Purification, partial characterization and anti-inflammatory characteristics of lotus (Nelumbo nucifera Gaertn) plumule polysaccharides. Food Chem. 2012; 135:1818–1827. PMID: 22953928.

8. Ahn JH, Kim ES, Lee C, Kim S, Cho SH, Hwang BY, Lee MK. Chemical constituents from Nelumbo nucifera leaves and their anti-obesity effects. Bioorg Med Chem Lett. 2013; 23:3604–3608. PMID: 23642481.

9. Mukherjee PK, Mukherjee D, Maji AK, Rai S, Heinrich M. The sacred lotus (Nelumbo nucifera) - phytochemical and therapeutic profile. J Pharm Pharmacol. 2009; 61:407–422. PMID: 19298686.
10. Sridhar KR, Bhat R. Lotus: a potential nutraceutical source. J Agric Technol. 2007; 3:143–155.
11. Bensky D, Clavey S, Stoger E. Chinese Herbal Medicine: Materia Medica. 3rd ed. Seattle (WA): Eastland Press;2015.
12. Li M, Xu Z. Quercetin in a lotus leaves extract may be responsible for antibacterial activity. Arch Pharm Res. 2008; 31:640–644. PMID: 18481022.

13. Yang D, Wang Q, Ke L, Jiang J, Ying T. Antioxidant activities of various extracts of lotus (Nelumbo nuficera Gaertn) rhizome. Asia Pac J Clin Nutr. 2007; 16(Suppl 1):158–163.
14. Lin HY, Kuo YH, Lin YL, Chiang W. Antioxidative effect and active components from leaves of Lotus (Nelumbo nucifera). J Agric Food Chem. 2009; 57:6623–6629. PMID: 19572539.

15. Mukherjee PK, Saha K, Das J, Pal M, Saha BP. Studies on the anti-inflammatory activity of rhizomes of Nelumbo nucifera. Planta Med. 1997; 63:367–369. PMID: 9270384.
16. Liu CP, Tsai WJ, Lin YL, Liao JF, Chen CF, Kuo YC. The extracts from Nelumbo Nucifera suppress cell cycle progression, cytokine genes expression, and cell proliferation in human peripheral blood mononuclear cells. Life Sci. 2004; 75:699–716. PMID: 15172179.

17. Ono Y, Hattori E, Fukaya Y, Imai S, Ohizumi Y. Anti-obesity effect of Nelumbo nucifera leaves extract in mice and rats. J Ethnopharmacol. 2006; 106:238–244. PMID: 16495025.

18. Tsuruta Y, Nagao K, Kai S, Tsuge K, Yoshimura T, Koganemaru K, Yanagita T. Polyphenolic extract of lotus root (edible rhizome of Nelumbo nucifera) alleviates hepatic steatosis in obese diabetic db/db mice. Lipids Health Dis. 2011; 10:202. PMID: 22067945.

19. Chen S, Fang L, Xi H, Guan L, Fang J, Liu Y, Wu B, Li S. Simultaneous qualitative assessment and quantitative analysis of flavonoids in various tissues of lotus (Nelumbo nucifera) using high performance liquid chromatography coupled with triple quad mass spectrometry. Anal Chim Acta. 2012; 724:127–135. PMID: 22483220.

20. Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. ScientificWorldJournal. 2013; 2013:162750. PMID: 24470791.

21. Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, Trethewey RN, Lange BM, Wurtele ES, Sumner LW. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004; 9:418–425. PMID: 15337491.

22. Huang B, Ban X, He J, Tong J, Tian J, Wang Y. Comparative analysis of essential oil components and antioxidant activity of extracts of Nelumbo nucifera from various areas of China. J Agric Food Chem. 2010; 58:441–448. PMID: 19919095.

23. Zhu MZ, Wu W, Jiao LL, Yang PF, Guo MQ. Analysis of flavonoids in lotus (Nelumbo nucifera) leaves and their antioxidant activity using macroporous resin chromatography coupled with LC-MS/MS and antioxidant biochemical assays. Molecules. 2015; 20:10553–10565. PMID: 26060918.

24. Naowaratwattana W, De-Eknamkul W, De Mejia EG. Phenolic-containing organic extracts of mulberry (Morus alba L.) leaves inhibit HepG2 hepatoma cells through G2/M phase arrest, induction of apoptosis, and inhibition of topoisomerase IIalpha activity. J Med Food. 2010; 13:1045–1056. PMID: 20828312.
25. Cuevas-Valenzuela J, González-Rojas A, Wisniak J, Apelblat A, Pérez-Correa JR. Solubility of (+)-catechin in water and water-ethanol mixtures within the temperature range 277.6-331.2 K: fundamental data to design polyphenol extraction processes. Fluid Phase Equilib. 2014; 382:279–285.
26. Auclair S, Milenkovic D, Besson C, Chauvet S, Gueux E, Morand C, Mazur A, Scalbert A. Catechin reduces atherosclerotic lesion development in apo E-deficient mice: a transcriptomic study. Atherosclerosis. 2009; 204:e21–e27. PMID: 19152914.

27. Mangiapane H, Thomson J, Salter A, Brown S, Bell GD, White DA. The inhibition of the oxidation of low density lipoprotein by (+)-catechin, a naturally occurring flavonoid. Biochem Pharmacol. 1992; 43:445–450. PMID: 1540202.

28. Weyant MJ, Carothers AM, Dannenberg AJ, Bertagnolli MM. (+)-Catechin inhibits intestinal tumor formation and suppresses focal adhesion kinase activation in the min/+ mouse. Cancer Res. 2001; 61:118–125. PMID: 11196148.
29. Handa O, Naito Y, Takagi T, Ishikawa T, Ueda M, Matsumoto N, Kokura S, Ichikawa H, Yoshida N, Shimoi K, Yoshikawa T. Inhibitory effects of catechins on neutrophil-dependent gastric inflammation. Redox Rep. 2002; 7:324–328. PMID: 12688521.

30. Iijima T, Mohri Y, Hattori Y, Kashima A, Kamo T, Hirota M, Kiyota H, Makabe H. Synthesis of (-)-epicatechin 3-(3-O-methylgallate) and (+)-catechin 3-(3-O-methylgallate), and their anti-inflammatory activity. Chem Biodivers. 2009; 6:520–526. PMID: 19353533.

31. Monga J, Aggarwal V, Suthar SK, Monika , Nongalleima K, Sharma M. Topical (+)-catechin emulsified gel prevents DMBA/TPA-induced squamous cell carcinoma of the skin by modulating antioxidants and inflammatory biomarkers in BALB/c mice. Food Funct. 2014; 5:3197–3207. PMID: 25310126.

32. Ademosun AO, Oboh G, Bello F, Ayeni PO. Antioxidative properties and effect of quercetin and its glycosylated form (Rutin) on acetylcholinesterase and butyrylcholinesterase activities. J Evid Based Complementary Altern Med. 2016; 21:NP11–NP17. PMID: 26438716.

33. Endale M, Park SC, Kim S, Kim SH, Yang Y, Cho JY, Rhee MH. Quercetin disrupts tyrosine-phosphorylated phosphatidylinositol 3-kinase and myeloid differentiation factor-88 association, and inhibits MAPK/AP-1 and IKK/NF-kappaB-induced inflammatory mediators production in RAW 264.7 cells. Immunobiology. 2013; 218:1452–1467. PMID: 23735482.
34. Maurya AK, Vinayak M. Anticarcinogenic action of quercetin by downregulation of phosphatidylinositol 3-kinase (PI3K) and protein kinase C (PKC) via induction of p53 in hepatocellular carcinoma (HepG2) cell line. Mol Biol Rep. 2015; 42:1419–1429. PMID: 26311153.

35. Ahn JY, Choi SE, Jeong MS, Park KH, Moon NJ, Joo SS, Lee CS, Choi YW, Li K, Lee MK, Lee MW, Seo SJ. Effect of taxifolin glycoside on atopic dermatitis-like skin lesions in NC/Nga mice. Phytother Res. 2010; 24:1071–1077. PMID: 20041431.

36. Ince I, Yesil-Celiktas O, Karabay-Yavasoglu NU, Elgin G. Effects of Pinus brutia bark extract and Pycnogenol in a rat model of carrageenan induced inflammation. Phytomedicine. 2009; 16:1101–1104. PMID: 19577447.

37. Kwon JH, Kim SB, Park KH, Lee MW. Antioxidative and anti-inflammatory effects of phenolic compounds from the roots of Ulmus macrocarpa. Arch Pharm Res. 2011; 34:1459–1466. PMID: 21975807.

38. Wang YH, Wang WY, Chang CC, Liou KT, Sung YJ, Liao JF, Chen CF, Chang S, Hou YC, Chou YC, Shen YC. Taxifolin ameliorates cerebral ischemia-reperfusion injury in rats through its anti-oxidative effect and modulation of NF-kappa B activation. J Biomed Sci. 2006; 13:127–141. PMID: 16283433.

39. Sung J, Lee J. Anti-inflammatory activity of butein and luteolin through suppression of NFκB activation and induction of Heme Oxygenase-1. J Med Food. 2015; 18:557–564. PMID: 25692285.

40. Lamy S, Moldovan PL, Ben Saad A, Annabi B. Biphasic effects of luteolin on interleukin-1β-induced cyclooxygenase-2 expression in glioblastoma cells. Biochim Biophys Acta. 2015; 1853:126–135. PMID: 25409926.

41. Nunes C, Almeida L, Barbosa RM, Laranjinha J. Luteolin suppresses the JAK/STAT pathway in a cellular model of intestinal inflammation. Food Funct. 2017; 8:387–396. PMID: 28067377.

Fig. 1
Effects of NLEs on the viability of RAW 264.7 cells, determined by MTT assay.
The cells were treated with NLEs for 24 h and then stimulated with LPS for 24 h. The values are expressed as the means ± SE of three individual experiments. LPS, lipopolysaccharide; NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract.
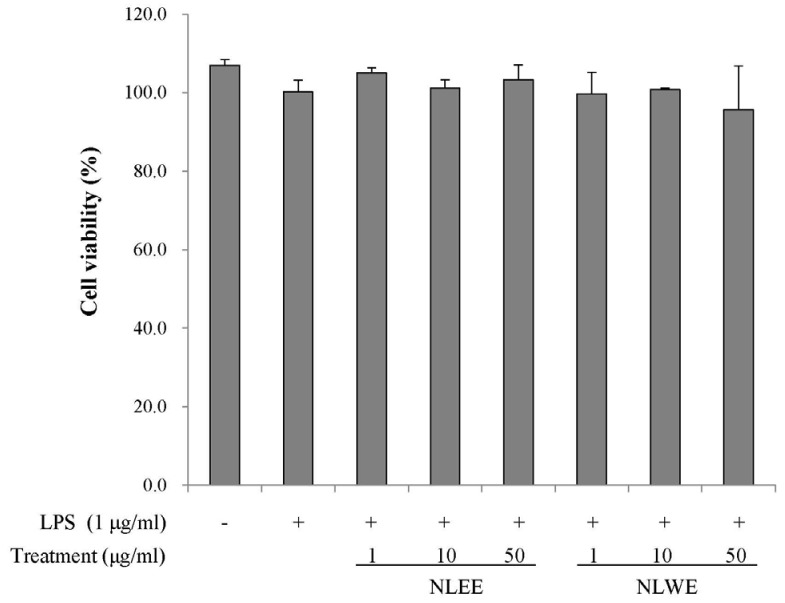
Fig. 2
Effects of NLEs on NO levels, and iNOS expression in LPS-induced RAW 264.7 cells.
The cells were pre-treated with various concentrations of NLE for 24 h, and then stimulated with LPS (1 µg/ml). (A) NO levels in cell culture media after 16-h incubation with LPS. (B) iNOS mRNA expression, determined by semi-quantitative RT-PCR after 4 h incubation with LPS. (C) iNOS protein expression in cell lysates, determined by western blot analysis. The values are expressed as the mean ± SD of three individual experiments. Values not sharing the same superscript were significantly (P < 0.05) different. LPS, lipopolysaccharide; NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract; NO, nitric oxide; iNOS, inducible nitric oxide synthase.
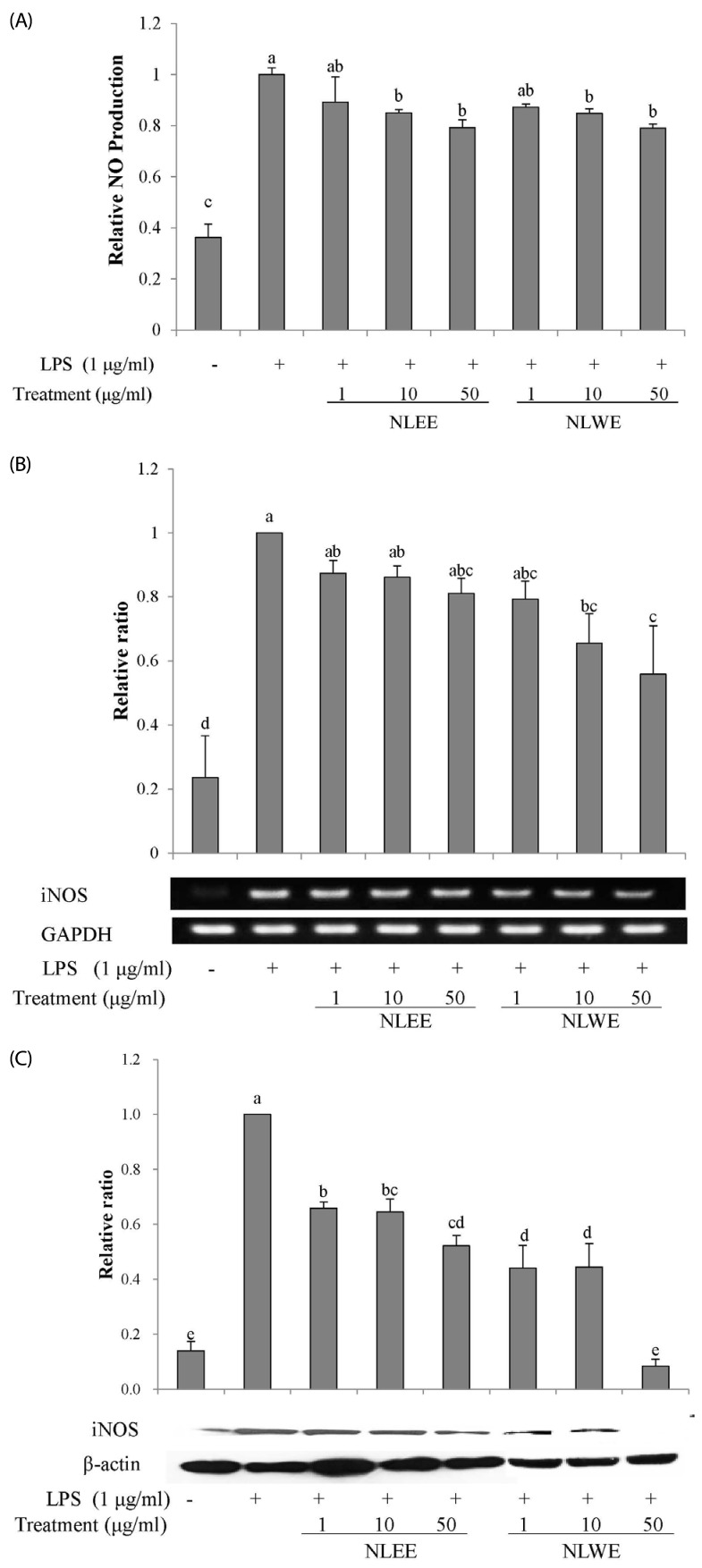
Fig. 3
Effects of NLEs on PGE2 levels and COX-2 expression in LPS-induced RAW 264.7 cells.
Cells were pre-treated with NLE for 24 h and then stimulated with LPS (1 µg/ml). (A) PGE2 levels were measured in the culture medium after 16 h incubation with LPS. (B) COX-2 mRNA expression was measured by semi-quantitative RT-PCR after 4 h incubation with LPS. (C) COX-2 protein levels in cell lysates were measured by western blot analysis after 16-h LPS stimulation. The values are expressed as the mean ± SE of three individual experiments. Values not sharing the same superscript were significantly (P < 0.05) different. LPS, lipopolysaccharide; NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract; PGE2, prostaglandin E2; COX-2, cyclooxygenase 2.
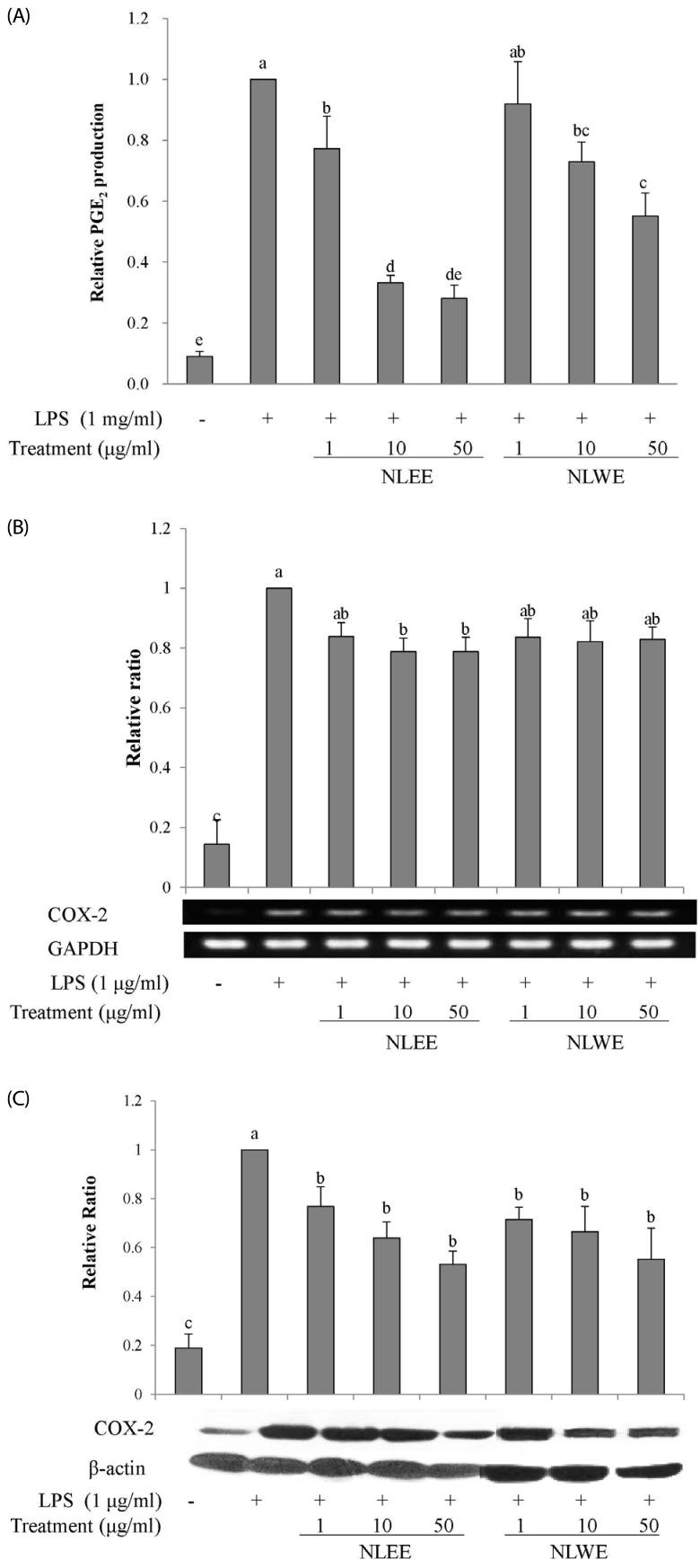
Fig. 4
Effects of NLEs on cytokine expression.
RAW 264.7 cells were pre-treated with NLEs for 24 h and then stimulated with LPS (1 µg/ml) for 4 or 16 h. Total RNA and protein lysates were subjected to semi-quantitative RT-PCR and western blot analysis to analyze IL-1β (A), IL-6 (B), and TNF-α (C). The values are expressed as the mean ± SD of three individual experiments. Values not sharing the same superscript were significantly (P < 0.05) different. LPS, lipopolysaccharide; NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract; IL, interleukin; TNF-α, tumor necrosis factor alpha; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
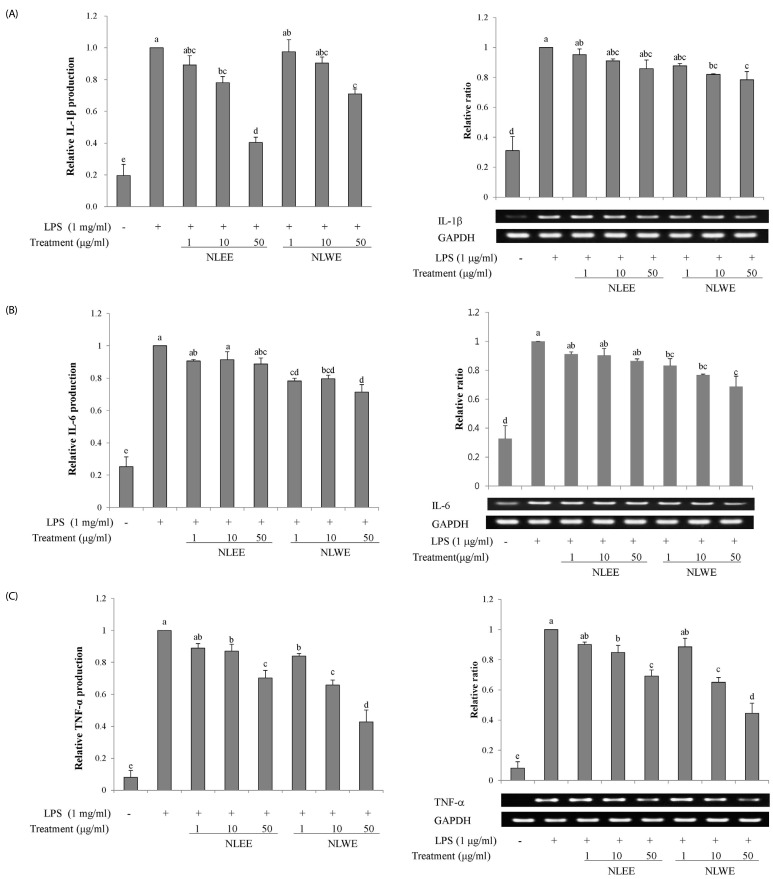
Fig. 5
Effects of NLE on NF-µB signaling.
(A) NF-µB transcriptional activity was determined in nuclear extracts of RAW 264.7 cells that were pre-treated with NLEs for 24 h prior to stimulation with LPS (1 µg/ml) for 20 min. (B) IµB phosphorylation was determined in cell lysate proteins from RAW 264.7 cells that were pre-treated with NLEs for 24 h and then induced with LPS (1 µg/ml) for 15 min. The values are expressed as the mean ± SD of three individual experiments. Values not sharing the same superscript were significantly (P < 0.05) different. LPS, lipopolysaccharide; NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract; NF-κB, nuclear factor kappa-light-chainenhancer of activated B cells; I-κB, inhibitor of NF-κB.

Fig. 6
PCA of the metabolites in NLWE (black circle) and NLEE (Red pentagon), analyzed by LC-MS/MS in (A) negative ion mode and (B) positive ion mode.
NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract.

Fig. 7
S-plot generated by OPLS-DA for the metabolites in NLWE and NLEE, analyzed by LC-ESI-ion trap-MS/MS.
(A) negative ion mode; (B) positive ion mode NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract.
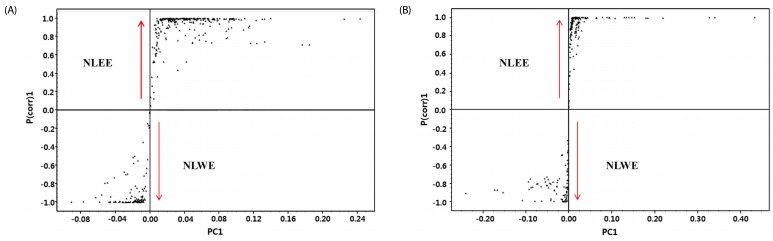
Fig. 8
Quantitative analysis of quercetin (A and B) and catechin (C and D) in NLWE and NLEE using HPLC.
Quercetin hydrate standard (A) and quercetin peak from NLEE and NLWE (B). Catechin hydrate standard (C) and catechin peak from NLEE and NLWE (D). NLEE, Nelumbo leaf ethanol extract; NLWE, Nelumbo leaf water extract.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download