Abstract
Purpose
Aberrant DNA methylation patterns have been commonly associated with human cancers. We have investigated the frequency of DNA hypermethylation in promoter regions from adenocarcinomas of the lung and then attempted to detect the same epigenetic changes from patient serum samples.
Materials and Methods
We collected tissues from 72 cases of lung adenocarcinomas. The cancer and normal lung tissues were tested for DNA hypermethylation using methylation-specific PCR (MSP). The genes investigated were DAPK, RARβ P2 and p16. We selected 12 patients where promoter hypermethylation was present for all three genes and four patients where hypermethylation was not seen for any of the three genes. Serum-free DNA was extracted and was tested for promoter hypermethylation. The status of serum-free DNA methylation was analyzed; the hypermethylation status was compared to clinical variables and cancer outcomes.
Results
DNA hypermethylation was observed in 32% of samples for DAPK, 63% of samples for RARβ P2 and 83% of samples for p16 from the cancer tissues. Among the 12 matched serum samples where the primary tumor showed hypermethylation in all three gene promoter regions, we were able to detect five incidences of serum DNA hypermethylation in four patients. The four patients had TNM stage II or higher disease. None of the patients with stage I disease showed serum-free DNA hypermethylation.
Conclusion
Aberrant promoter hypermethylation was frequently observed in surgically resected adenocarcinoma of the lung. Concurrent serum-free DNA hypermethylation was detected in 34% of patients where the primary tumor showed hypermethylation in all three gene promoter regions. The findings suggest that the serum-free DNA methylation status might be used as a potential target for the diagnosis of lung cancer. However, the low sensitivity should be improved for use in a clinical application.
Go to : 
References
1. Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1999. CA Cancer J Clin. 1999; 49:8–31.

2. Bunn PJ Jr. Early detection of lung cancer using serum RNA or DNA markers: ready for "prime time" or for validation? J Clin Oncol. 2003; 21:3891–3893.

3. Kim YT, Lee SH, Sung SW, Kim JH. Can aberrant promoter hypermethylation of CpG islands predict the clinical outcome of nonsmall cell lung cancer after curative resection? Ann Thorac Surg. 2005; 79:1180–1188.

4. Kim YT, Park SJ, Lee SH, et al. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005; 130:1378.

5. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61:1659–1665.
6. Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003; 21:3902–3908.

7. Sozzi G, Conte D, Mariani L, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001; 61:4675–4678.
8. Bearzatto A, Conte D, Frattini M, et al. p16 (INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from nonsmall cell lung cancer. Clin Cancer Res. 2002; 8:3782–3787.
9. Beau-Faller M, Gaub MP, Schneider A, et al. Plasma DNA microsatellite panel as sensitive and tumor-specific marker in lung cancer patients. Int J Cancer. 2003; 105:361–370.

10. Gormally E, Hainaut P, Caboux E, et al. Amount of DNA in plasma and cancer risk: a prospective study. Int J Cancer. 2004; 111:746–749.

11. Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic Acid as prognostic marker in nonsmall-cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004; 22:4157–4164.

12. Herrera LJ, Raja S, Gooding WE, et al. Quantitative analysis of circulating plasma DNA as a tumor marker in thoracic malignancies. Clin Chem. 2005; 51:113–118.

13. Chen XQ, Stroun M, Magnenat JL, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996; 2:1033–1035.

14. Sozzi G, Musso K, Ratcliffe C, Goldstraw P, Pierotti MA, Pastorino U. Detection of microsatellite alterations in plasma DNA of nonsmall cell lung cancer patients: a prospect for early diagnosis. Clin Cancer Res. 1999; 5:2689–2692.
15. Andriani F, Conte D, Mastrangelo T, et al. Detecting lung cancer in plasma with the use of multiple genetic markers. Int J Cancer. 2004; 108:91–96.

16. Cuda G, Gallelli A, Nistico A, et al. Detection of microsatellite instability and loss of heterozygosity in serum DNA of small and nonsmall cell lung cancer patients: a tool for early diagnosis? Lung Cancer. 2000; 30:211–214.

17. Ramirez JL, Sarries C, de Castro PL, et al. Methylation patterns and K-ras mutations in tumor and paired serum of resected nonsmall-cell lung cancer patients. Cancer Lett. 2003; 193:207–216.

18. Kimura T, Holland WS, Kawaguchi T, et al. Mutant DNA in plasma of lung cancer patients: potential for monitoring response to therapy. Ann N Y Acad Sci. 2004; 1022:55–60.

19. Silva JM, Gonzalez R, Dominguez G, Garcia JM, Espana P, Bonilla F. TP53 gene mutations in plasma DNA of cancer patients. Genes Chromosomes Cancer. 1999; 24:160–161.

20. An Q, Liu Y, Gao Y, et al. Detection of p16 hypermethylation in circulating plasma DNA of nonsmall cell lung cancer patients. Cancer Lett. 2002; 188:109–114.

21. Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from nonsmall cell lung cancer patients. Cancer Res. 1999; 59:67–70.
22. Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002; 62:371–375.
Go to : 
Figures and Tables
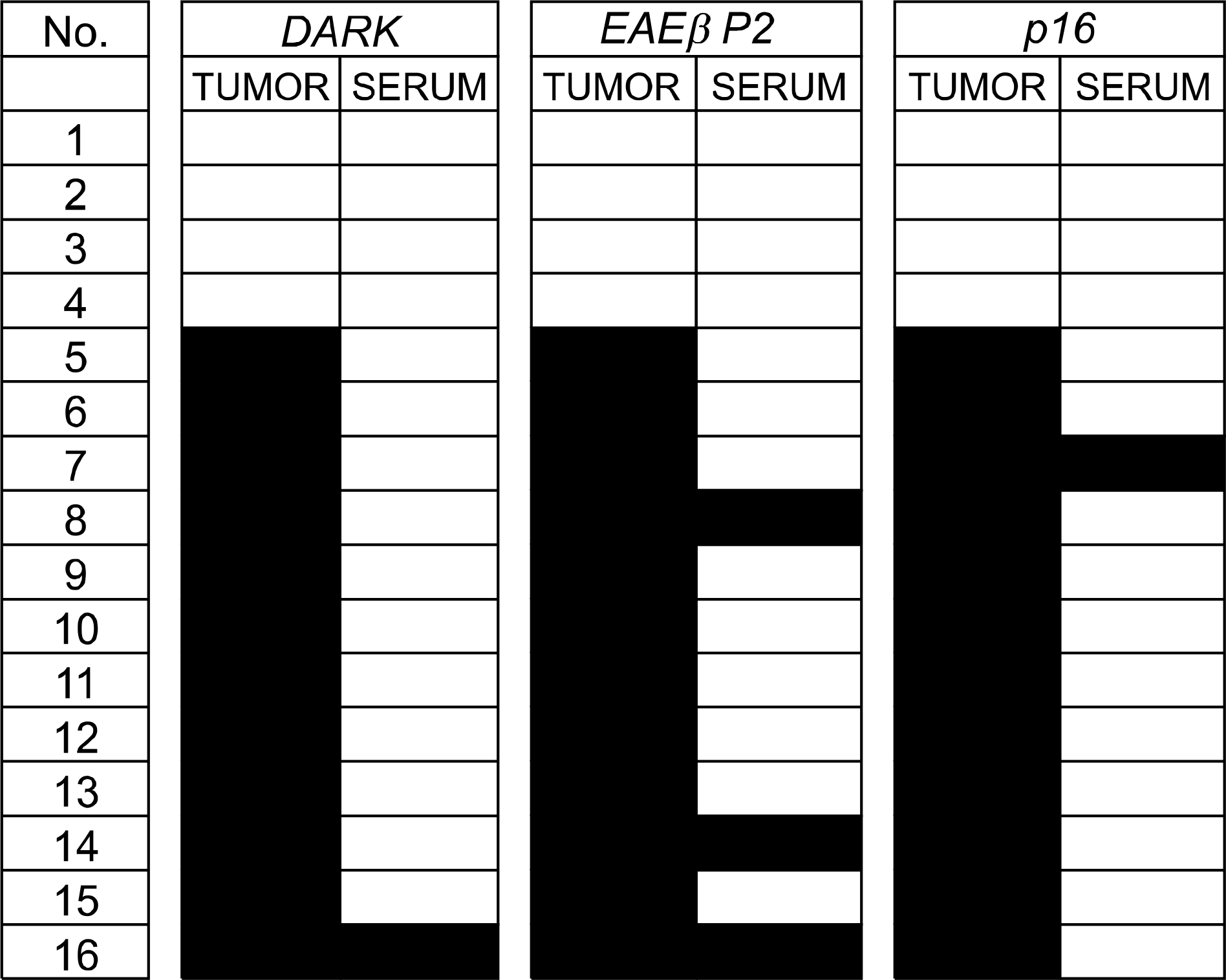 | Fig. 2.Summary of the methylation status for DAPK, RARβ P2 and p16 in primary tumor and serum samples. The black-colored boxes represent methylated samples. |
Table 1.
Summary of Primer Sequences, Annealing Temperatures and PCR Product Sizes Used for MSP
Table 2.
Methylation Frequency of the Cancer Tissues According to the TNM Stage
| Gene | Early stage | Advanced stage | p value* | ||||
|---|---|---|---|---|---|---|---|
| Stage Ia (13) | Stage Ib (19) | Stage IIb (11) | Stage IIIa (22) | Stage IIIb (6) | Stage IV (1) | ||
| p16 | 11 (84.6%) | 17 (89.5%) | 8 (72.7%) | 18 (81.8%) | 5 (83.3%) | 1 (100.0%) | 0.914 |
| RARβ P2 | 10 (76.9%) | 12 (63.2%) | 6 (54.5%) | 12 (54.5%) | 4 (66.7%) | 1 (100.0%) | 0.577 |
| DAPK | 3 (23.1%) | 7 (36.8%) | 4 (36.4%) | 8 (36.4%) | 2 (33.3%) | 0 (0.0%) | 0.865 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download