Abstract
Purpose:
Our aim of research is to find novel genes that overexpressed in various samples such as cell tines and tissues that infinitely proliferate; so-called immortalized cells, cancer cells and tissues. In this study, we obtained a gene from immortalized cell line (WI-38 VA13) then identified it from various cell lines and human lung tissues.
Materials and Methods:
Using suppressive subtractive hybridization (SSH) method, we obtained many genes and selected a novel gene of them. And then the novel gene fragment was amplified by PCR and ligated in Τ easy vector for sequencing. And finally we found a differentially expressed gene in cell lines and tissues when it was performed by reverse transcriptase-PCR (RT-PCR),
Results:
As the result of transformation of genes that we gained using SSH, we obtained about 150 clones. And then we certificated several genes by DNA prep and confirmed it by sequencing. We examined whether the gene sequences are novel or known genes by genome homology search and we confirmed the gene expressions by RT-PCR. As a result, we identified a differentially overexpressed gene (named “clone 58”) in immortalized cells, cancer cell lines and lung squamous cell carcinomas.
Conclusion:
The “clone 58” mRNA was significantly up-regulated in various human cell lines and also human lung cancer tissues compared to the normal. We suppose that this gene can carry out a specific role related to the induction of cancer and/or the mechanism of the changeover of a normal cell to an immortalized cell. In short, the discovery of our gene has an importance in the point that they are thought to have a connection with immortalization and carcinogenesis of human cells and tissues, (J Lung Cancer 2006;5(2):96 — 101)
Go to : 
REFERENCES
1.National Institutes of Health. National Cancer Institute. Recognizing symptoms. Available at: http://www.nci.nih.gov/cancer-topics/wyntk/lung/page6. Accessed December 12:. 2004.
2.Lilienfeld DE., Mandel JS: Coin P: Schuman LM. Projection of asbestos related diseases in the United States, 1985–2009. Br J Ind Med. 1988. 45:283–29. L.
3.Fabricius P., Lange P. Diet and lung cancer. Monaldi Archives of Chest Diseases. Monaldi Arch Chest Dis. 2003. 59:207–211.
4.Ferri FF. Carcinoma of the lung. Ferri FF, editor. editor.Practical Guide of the Care of the Medical Patient. 6th ed.St. Louis, MO; Mosby, Inc;2004. p. 507–508.
5.Sasaki M., Sugio K., Kuwabara Y, et al. Alterations of tumor suppressor genes (Rb, pi6: p27 and p53) and an increased FDG uptake in lung cancer. Ann Nucl Med. 2003. 17:189–196.
6.Ludlow JW. Interaction between SV40 large-tumor antigen and the growth suppressor pRB and p53. FASEB J. 1993. 7:866–871.
7.Woenckhaus M., Klein-Hitpass L., Grepmeier U, et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers, J Pathol. 2006. 210:192–204.
8.Hufton SE., Moerkerk PT: Brandwijk R: de Bruine AP., Arends JW: Hoogenboom HR. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 1999. 463:77–82.

9.Ran Yuliang., Jiang Yangfu., Xing Zhong, et al. Identification of derlin-1 as a novel growth factor-responsive endothelial antigen by suppression subtractive hybridization. BBRC. 2006. 348:1272–1278.

10.He N: Qin Q: Xu X. Differential profile of genes expressed in gemocytes of White Spot Syndrome Virus-resistant shrimp (Penaeus japonicus) by combining suppression subtractive hybridization and differential hybridization. Antiviral Res. 2005. 66:39–45.
11.Sallinen SL: Sallinen PK: Haapasalo HK, et al. identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000. 60:6617–6622.
12.Liu YB., Wei ZX: Li L: Li HS: Chen H., Li XW. Construction and analysis of SSH cDNA library of human vascular endothelial cells related to gastrocarcinoma. World J Gastroenterol. 2003. 9:19–23.

13.Shridhar V: Sen A., Chien J, et al. Identification of underexpressed genes in early and late-stage primary ovarian tumors by wuppression subtraction hybridization. Cancer Res. 2002. 62:262–270.
14.Sekido Y: Fong KM., Minna JD. Epidemiology of lung cancer. In:. Murray JF: Nadel JA, editor. ediorts.Textbook of Respiratory Medicine. 3rd ed.Philadelphia, PA; W.B. Saunders Company;2000. p. 1397–1406.
15.Osann KE., Lowery JT: Schell MJ. Biology of lung cancer. In: Murray JF: Nadel JA: editors. Textbook of Respiratory Medicine. 3rd ed.Philadelphia, PA; W.B. Saunders Company;2000. p. 1375–1379.
16.US Department of Health and Human Services. The health consequences of involuntary smoking. A Report of the Surgeon General. Publication No. (CDC)87-8398. Rockville: MD: Department of Health and Human Services, Office on Smoking and Health. 1987.
17.Kim JY: Chung YS., Faek KH, et al. Isolation and characterization of a cDNA encoding the cysteine proteinase inhibitor, induced upon flower maturation in carnation using suppression subtractive hybridization. Mol Cells. 1999. 9:392–397.
18.Liang P., Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992. 257:967–971.

19.Vasmatzis G., Essand M., Brinkmann U., Lee B., Pastan I. Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc Natl Acad Sci USA. 1998. 95:300–304.

20.Porkka KP., Visakorpi T. Detection of differentially expressed genes in prostate cancer by combining suppression subtractive hybridization and cDNA library array. J Pathol. 2001. 193:73.

21.McClelland M., Chanda K: Welsh J., Ralph D. Arbitrary primed PCR fingerpriming of RNA applied to mapping differentially expressed genes. EXS. 1993. 67:103–115.
22.Dong Y., Zhang H: Hawthorn L: Ganther HE., Ip C. Delineation of the molecular basis for selenium-induced growth arrest in human prostate cancer cells by oligonucleotide array. Cancer Res. 2003. 63:52–59.
23.Cheng J., Imanishi H., Liu W, et al. Involvement of cell cycle regulatory proteins and MAP kimase signaling pathway in growth inhibition and cell cycle arrest by a selective cyclooxy-genase2 carcinoma cell lines. Cancer Sci. 2004. 95:666–673.
Go to : 
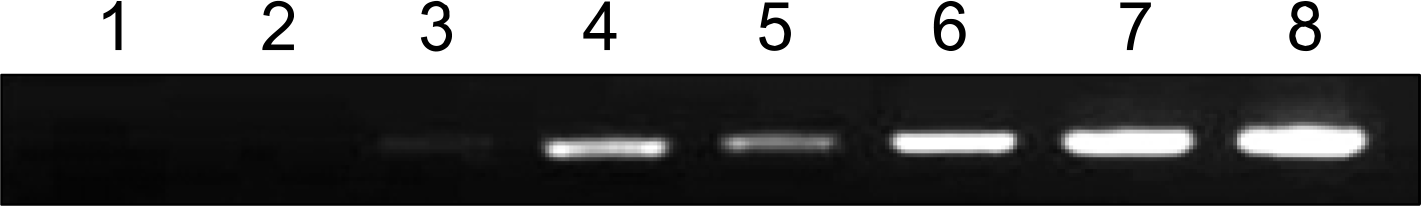 | Fig. 1.Reduction of G3PDH abundance by PCR-select subtraction. Normal cDNA was prepared from WI38 (human lung fibroblast cell) total RNA and immortalization cDNA was prepared from WI38 VA13 (transformed WI38 by SV40), PCR was performed on subtracted (lanes 1 〜4) or unsubtracted (lanes 5-8) secondary PCR product with the G3PDH 5’ and 3’ primers. Lanes 1, 5- 18 cycleså lanes 2ô6: 23 cycles; lanes 3, 7’ 28 cycles, lanes 4, 8: 33 cycles. |
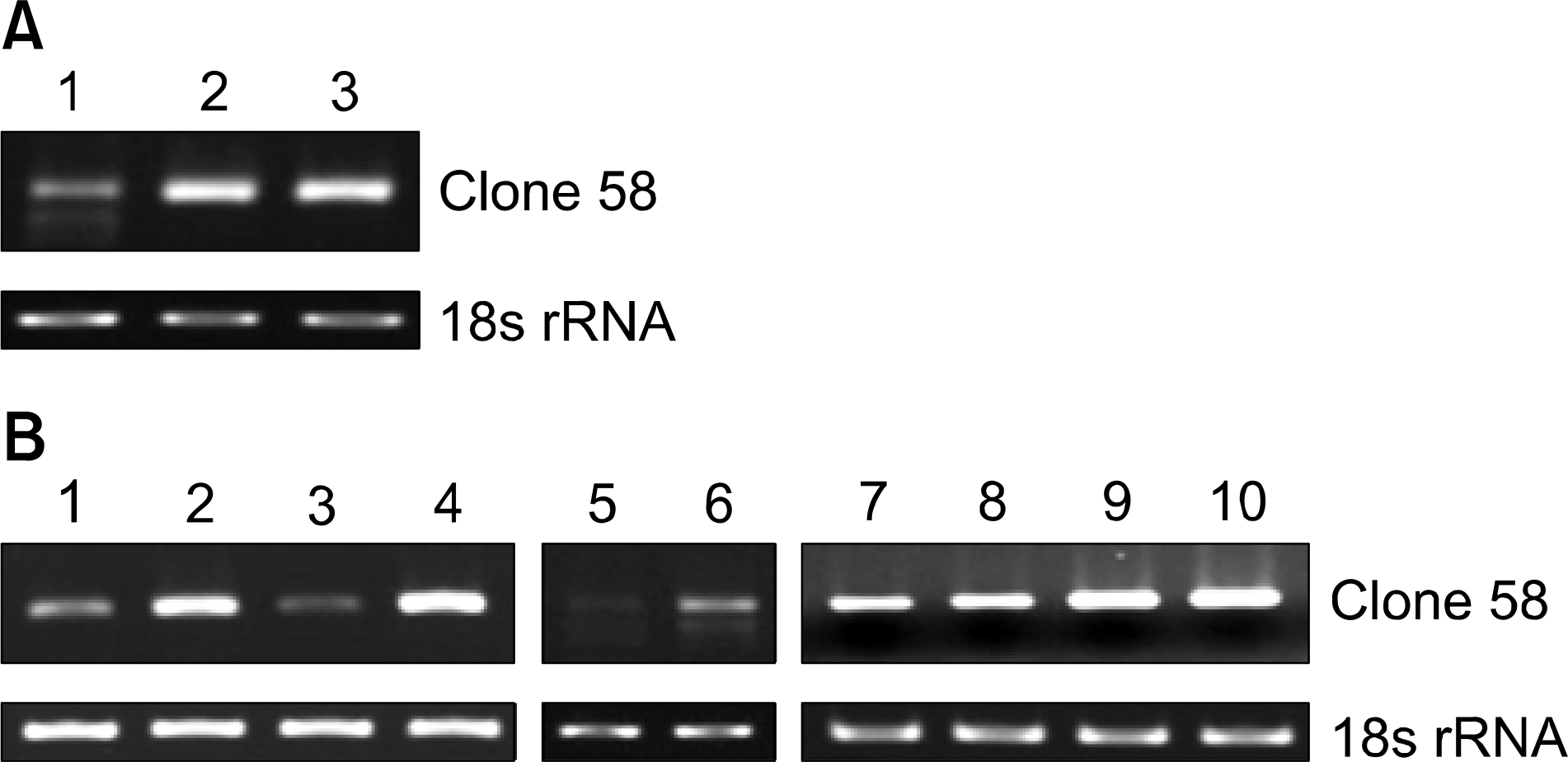 | Fig. 3.Expression of clone 58 in Wᅵ38ôWᅵ38 VA13 (A) and various cell lines (B), (A) Lane 1æ WI38 (lung fibroblast normal cell), lane 2æ WI38 VA13 (SV-40 transfected cell), lane 3æ WI26VA4 (SV-40 transfected cell). (B) Lane 1- skin normal cell, lane 2’ skin tumor cell, lane 3: colon normal cell, lane 4: colon tumor cell, lane 5- prostate normal cell, lane 6: prostate tumor cell, lane 7: SK (SK- N-SH): brain neuroblastoma, metastasis to bone marrow, lane 8: A172: brain, glioblastoma, lane 9: HeLa, lane 10æ Lung cancer. 18 s rRNA was used as the reference gene. |
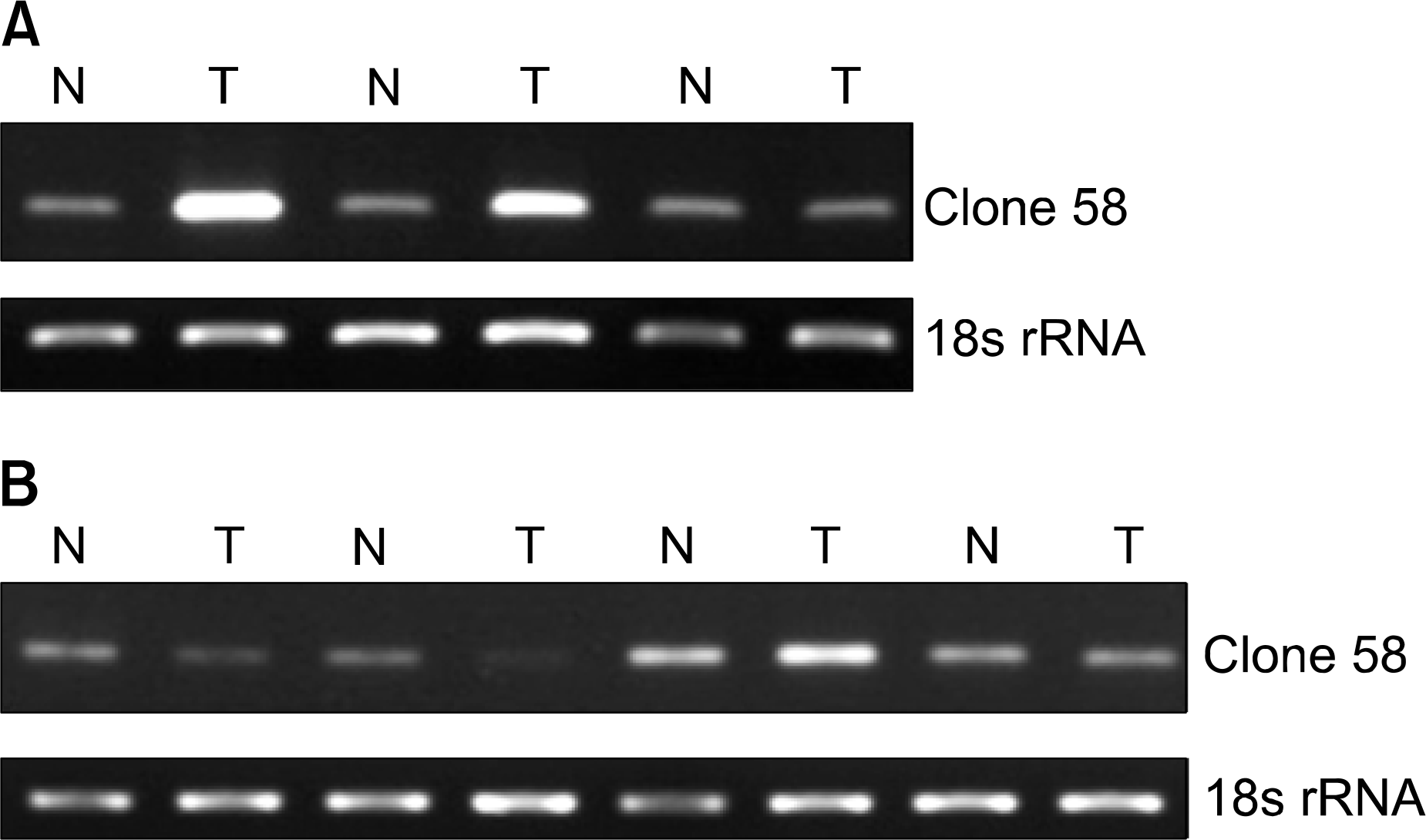 | Fig. 4.Clone 58 expression in non-small cell lung carcinoma tissues. (A) Næ normal lung tissues J: squamous cell carcinoma. (B) Næ normal lung tissues, T: adenocarcinoma 18s rRNA was used as the reference gene. |
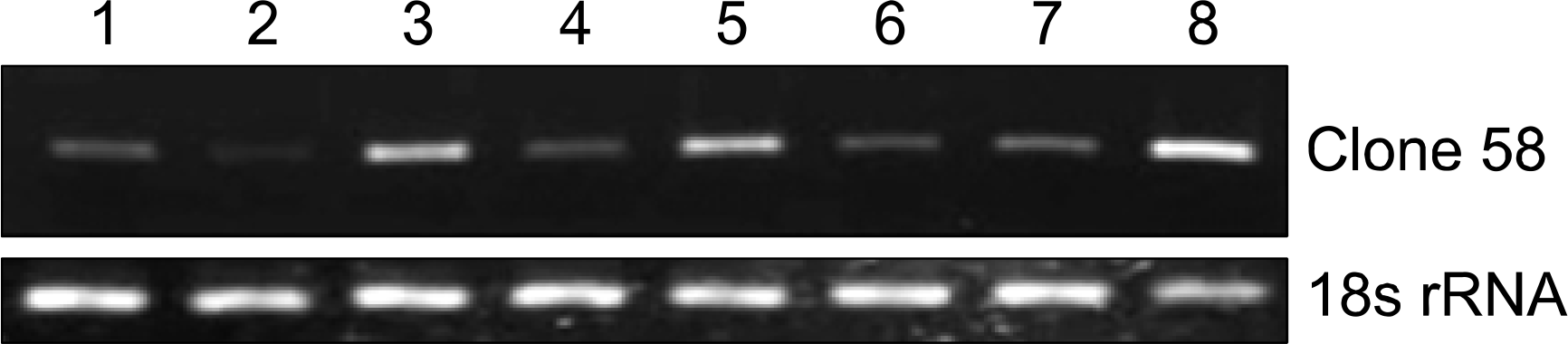 | Fig. 5.Serum siimulatioa Clone 58 expressions have no significant on time dependent manner. Lung fibroblast cell line (WI38) was used Lane 1: arrest stage, lane 2æ 0.5 hr, lane 3: 1 hr, lane 4: 2 hr, lane 5: 4 hr, lane 6: 8 hr, lane 7æ 12 hr, lane 8: 24 hr after serum stimulation. 18 s rRNA was used as the reference gene. |
Table 1.
Identification of Known Tumor Specific Genes from SSH Libraries




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download