Abstract
Purpose:
We applied a simplified method using polymerase chain reaction (PCR)-based enzymatic digestion for the detection of epidermal growth factor receptor ( EGFR) mutation. Materials and Methods: We selected 74 samples of adenocarcinoma of the lung with EGFR exons 19 and 21 that had been previously sequenced. We designed PCR primers and chose a DNA restriction enzyme. Seventy four additional lung cancer samples were tested as a test set. For test sets, the PCR-based method was performed first, followed by validation of the result by DNA sequencing.
Results:
In the first sample group, we found 15 (20.3%) mutations in exon 19, and 9 (12.2%) mutations in exon 21 using the sequencing method. By using the PCR-based method, we were able to identify all of the mutated samples detected by the sequencing method. The PCR-based method also detected mutations in exon 19 in three additional samples and in exon 21 in one additional sample. In the second sample group, by performing the PCR-based method, we found 10 (13.5%) and 7 (9.5%) mutations in exons 19 and 21, respectively. Additional mutations in exon 19 were identified in 2 samples by the sequencing method. However, the sequencing method failed to identify a mutation in exon 21 in one sample.
REFERENCES
1. Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005; 353:123–132.

2. Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004; 22:1103–1109.

3. Karamouzis MV, Grandis JR, Argiris A. Therapies directed against epidermal growth factor receptor in aerodigestive car-cinomas. JAMA. 2007; 298:70–82.

4. Paez JG, Jä nne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004; 304:1497–1500.
5. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004; 350:2129–2139.

6. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007; 7:169–181.

7. Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction- based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005; 7:396–403.
8. Janne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006; 12(3 Pt 1):751–758.

9. Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006; 354:526–528.
10. Nomoto K, Tsuta K, Takano T, et al. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006; 126:608–615.
11. Willmore-Payne C, Holden JA, Layfield LJ. Detection of epidermal growth factor receptor and human epidermal growth factor receptor 2 activating mutations in lung adenocarcinoma by high-resolution melting amplicon analysis: correlation with gene copy number, protein expression, and hormone receptor expression. Hum Pathol. 2006; 37:755–763.

12. Sasaki H, Endo K, Konishi A, et al. EGFR mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin Cancer Res. 2005; 11:2924–2929.
13. Asano H, Toyooka S, Tokumo M, et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res. 2006; 12:43–48.
14. Kimura H, Fujiwara Y, Sone T, et al. High sensitivity detection of epidermal growth factor receptor mutations in the pleural effusion of non-small cell lung cancer patients. Cancer Sci. 2006; 97:642–648.

15. Nagai Y, Miyazawa H, Huqun , et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005; 65:7276–7282.

16. Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006; 12(3 Pt 1):839–844.

17. Huang SF, Liu HP, Li LH, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004; 10:8195–8203.

18. Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004; 101:13306–13311.
Fig. 1.
Gel electrophoresis result of polymerase chain reaction-based method. Arrows indicate mutation band.
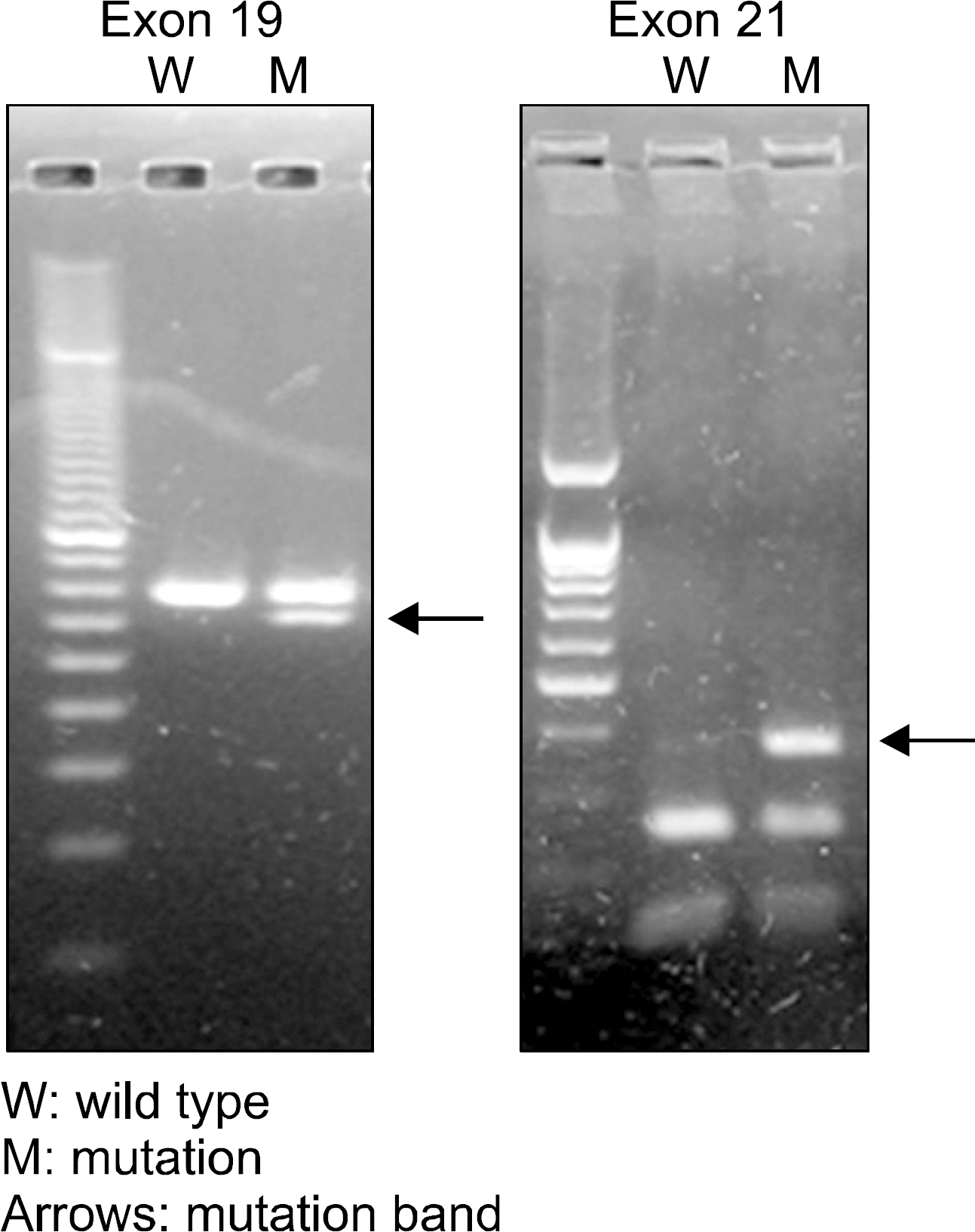
Fig. 2.
Re-testing result of two false negative cases of polymerase chain reaction-based method in training group for exon 19 mutations. Note faint bands underneath to the wild type DNA bands.
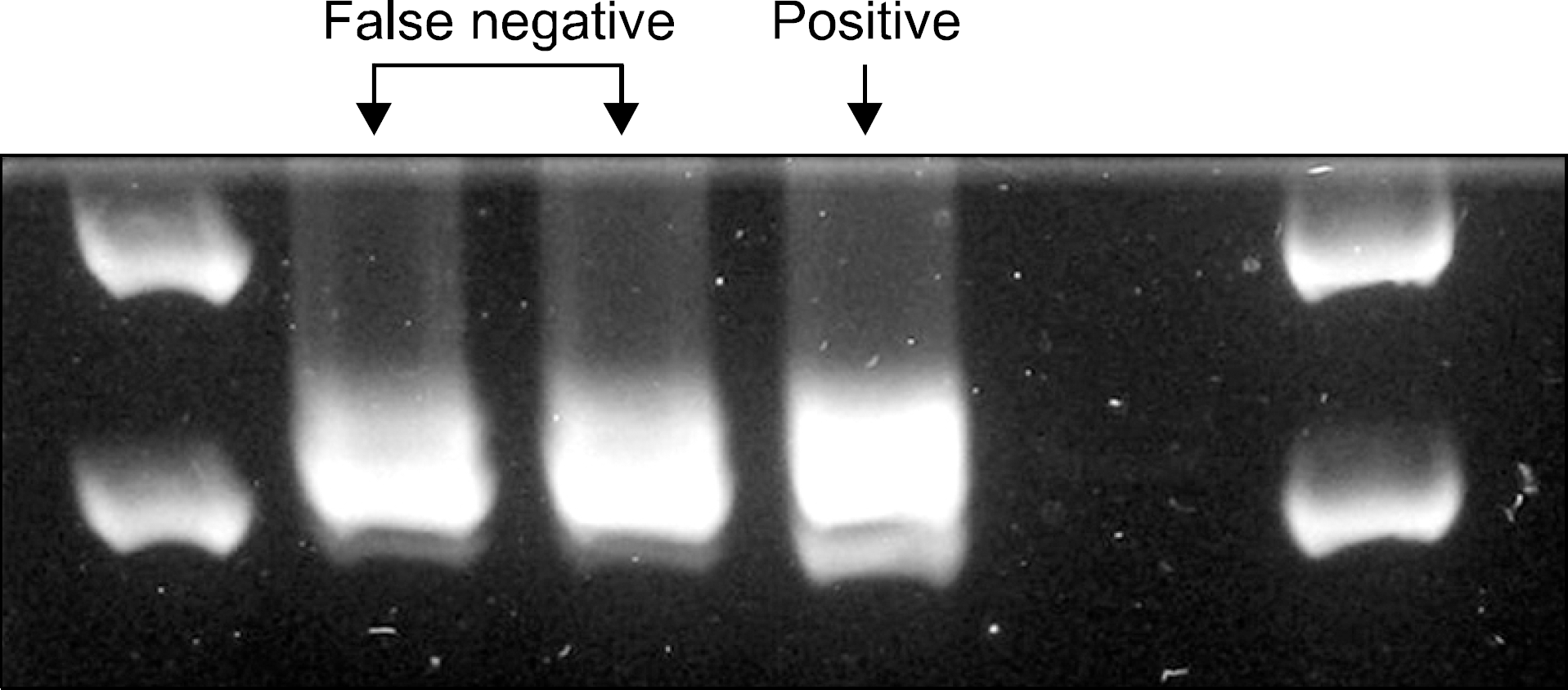
Table 1.
EGFR Mutations in Exons 18∼21 Tested by DNA Sequencing in the Training Set
Table 2.
Comparison of EGFR Mutation Detection Results Using the Sequencing and PCR-Based Methods in the Training Set
Table 3.
Comparison of EGFR Mutation Detection Results Using the Sequencing and PCR-Based Methods in the Test Set




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download