Abstract
Purpose:
The anaplastic lymphoma kinase ( ALK) gene is a potential molecular target in non-small cell lung carcinoma (NSCLC). The clinicopathologic implication of a change in the ALK gene copy number (GCN) is unclear. Materials and Methods: A total of 434 primary NSCLC samples were analyzed by fluorescence in situ hybridization (FISH) for ALK GCN.
Results:
Ninety-six cases (22.1%) showed ALK GCN gain with amplification in 16 (3.7%) cases. The cases with ALK GCN gain consisted of 47 adenocarcinomas (49.0%), 41 squamous cell carcinomas (42.7%), 5 adenosquamous carcinomas (5.2%) and 3 other NSCLCs (3.1%). ALK gene amplification was identified in 7 adenocarcinomas (43.7%) and 9 squamous cell carcinomas (56.3%). There was no significant difference between ALK GCN gain/amplification and histologic subtypes. Uni-variate survival analysis revealed that patients with ALK GCN gain/amplification showed shorter progression-free survival durations and decreased overall survival rates (p<0.001). However, multivariate analysis proved that ALK GCN gain/amplification is not an independent prognostic factor for progression-free survival or overall survival.
Go to : 
REFERENCES
2. Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002; 346:92–98.

3. Scagliotti GV, De Marinis F, Rinaldi M, et al. Phase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancer. J Clin Oncol. 2002; 20:4285–4291.

4. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007; 448:561–566.

5. Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010; 363:1693–1703.

6. Ou SH, Bazhenova L, Camidge DR, et al. Rapid and dramatic radiographic and clinical response to an ALK inhibitor (crizotinib, PF02341066) in an ALK translocation-positive patient with non-small cell lung cancer. J Thorac Oncol. 2010; 5:2044–2046.

7. Cools J, Wlodarska I, Somers R, et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibro-blastic tumor. Genes Chromosomes Cancer. 2002; 34:354–362.

8. Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008; 455:971–974.

9. Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009; 115:1723–1733.
10. Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical fea-tures and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009; 27:4247–4253.

11. Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010; 16:5581–5590.

12. Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011; 6:466–472.

Go to : 
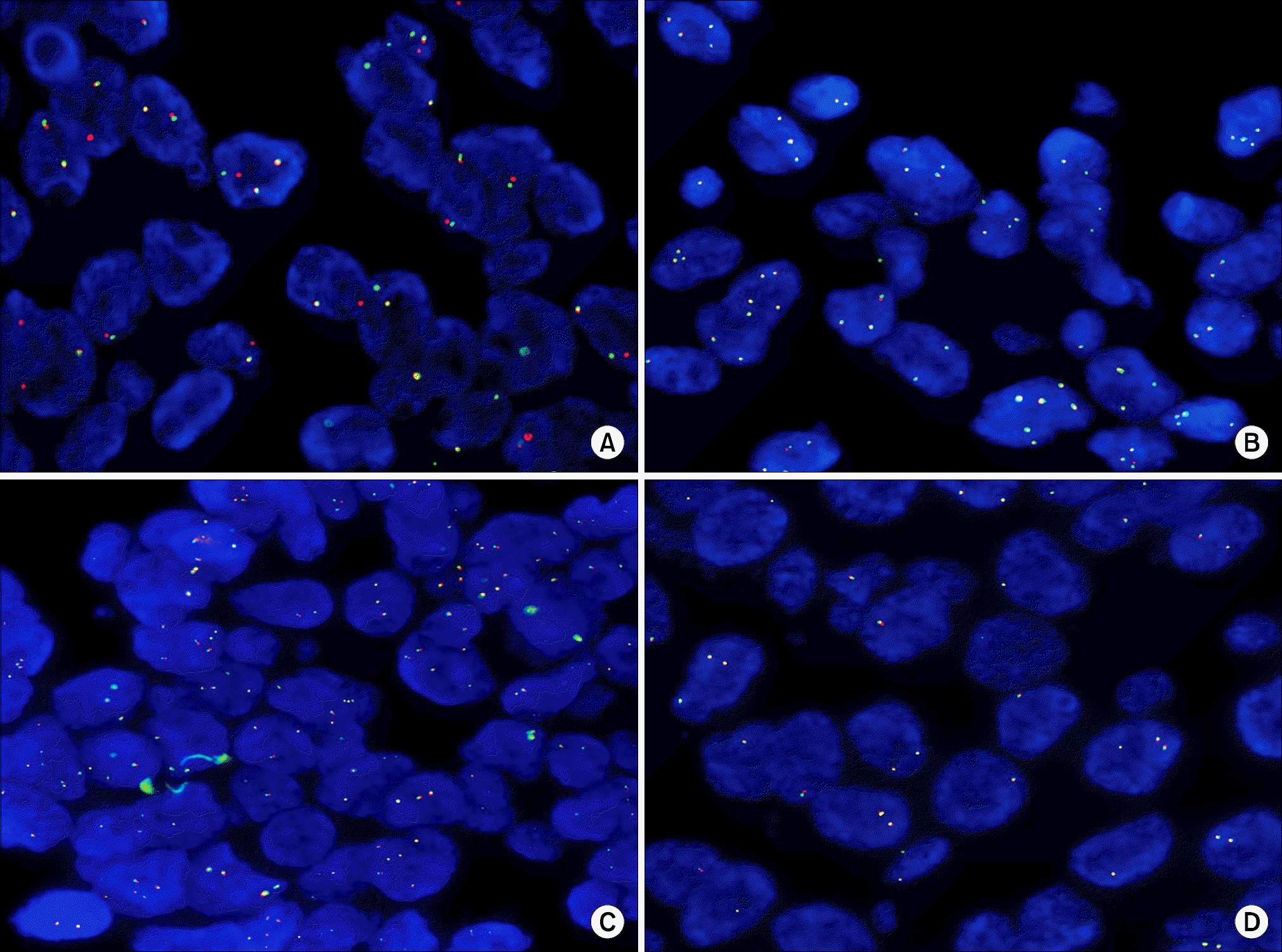 | Fig. 1.
ALK gene status by fluorescence in situ hybridization (FISH) (×1,000). (A) ALK gene rearrangement (break-apart pattern). (B) ALK gene copy number gain. (C) ALK gene amplification. (D) ALK wild type. |
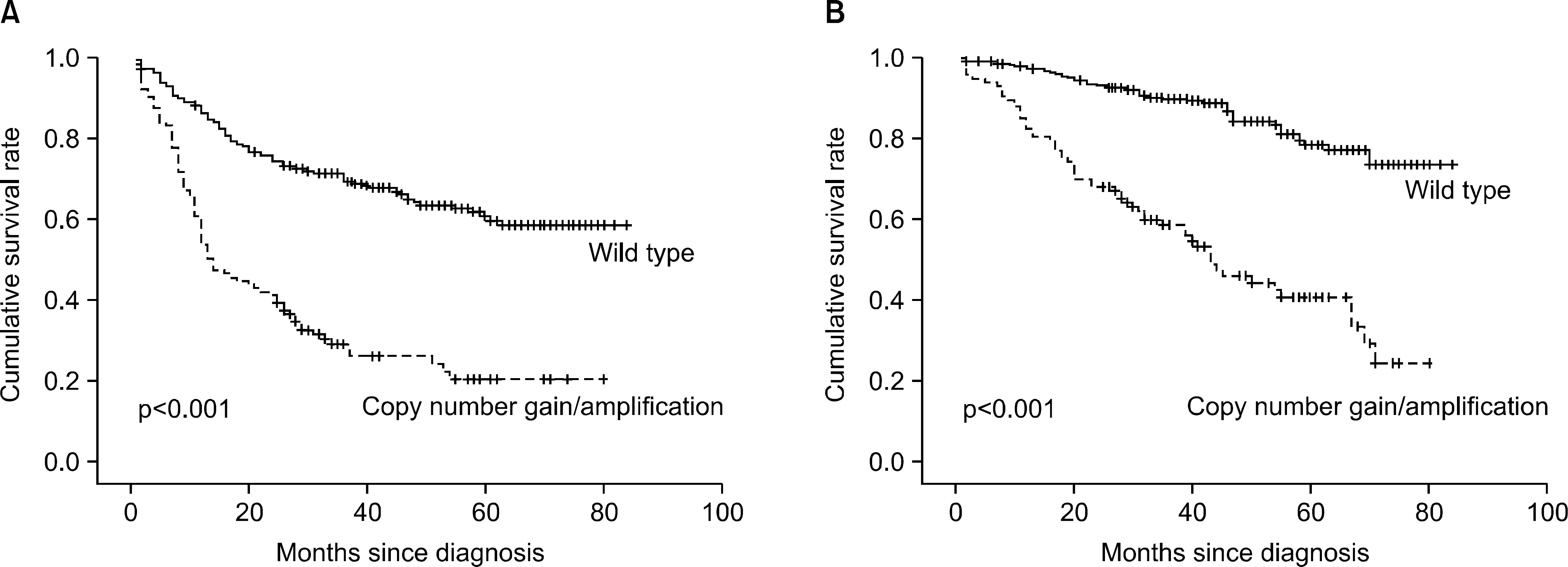 | Fig. 2.
ALK gene copy number gain/amplification showed significantly shorter progression-free survival (A) and overall-survival (B). |
Table 1.
Clinicopathologic Characteristics in ALK Gene Status
Table 2.
Clinicopathologic Characteristics in ALK Gene Status
Table 3.
Multivariate Analysis of Progression-free Survival




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download