Abstract
Microarray comparative genomic hybridization (CGH) has proven to be a specific, sensitive, and rapid technique, with considerable advantages compared to other methods used for analysis of DNA copy number changes. Array CGH allows for the mapping of genomic copy number alterations at the sub-microspecific level, thereby directly linking disease phenotypes to gene dosage alterations. The whole human genome can be scanned for deletions and duplications at over 30,000 loci simultaneously by array CGH (∼40 kb resolution). Array CGH can be used for analysis of DNA copy number aberrations that cause not only cancer and human genetic disease, but also normal human variation. This review gives the various array CGH platforms and their applications in cancer and human genetics. (J Lung Cancer 2011;10(2):77 ? 86)
Go to : 
REFERENCES
1. Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992; 258:818–821.

2. Weiss MM, Hermsen MA, Meijer GA, et al. Comparative genomic hybridisation. Mol Pathol. 1999; 52:243–251.

3. Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997; 13:405–409.

4. Trask BJ. Human cytogenetics: 46 chromosomes, 46 years and counting. Nat Rev Genet. 2002; 3:769–778.

5. Weiss MM, Kuipers EJ, Meuwissen SG, van Diest PJ, Meijer GA. Comparative genomic hybridisation as a supportive tool in diagnostic pathology. J Clin Pathol. 2003; 56:522–527.

6. Weiss MM, Kuipers EJ, Postma C, et al. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene. 2003; 22:1872–1879.

7. Forster-Gibson CJ, Davies J, MacKenzie JJ, Harrison K. Cryptic duplication of 21q in an individual with a clinical diagnosis of Down syndrome. Clin Genet. 2001; 59:438–443.

8. Levy B, Dunn TM, Kern JH, Hirschhorn K, Kardon NB. Delineation of the dup5q phenotype by molecular cytogenetic analysis in a patient with dup5q/del 5p (cri du chat). Am J Med Genet. 2002; 108:192–197.

9. Kallioniemi OP, Kallioniemi A, Piper J, et al. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994; 10:231–243.

10. Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet. 2004; 66:488–495.

11. Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998; 20:207–211.

12. Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999; 23:41–46.

13. Solinas-Toldo S, Lampel S, Stilgenbauer S, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997; 20:399–407.

14. Garnis C, Baldwin C, Zhang L, Rosin MP, Lam WL. Use of complete coverage array comparative genomic hybridization to define copy number alterations on chromosome 3p in oral squamous cell carcinomas. Cancer Res. 2003; 63:8582–8585.
15. Coe BP, Henderson LJ, Garnis C, et al. High-resolution chromosome arm 5p array CGH analysis of small cell lung carcinoma cell lines. Genes Chromosomes Cancer. 2005; 42:308–313.

16. Henderson LJ, Coe BP, Lee EH, et al. Genomic and gene expression profiling of minute alterations of chromosome arm 1p in small-cell lung carcinoma cells. Br J Cancer. 2005; 92:1553–1560.

17. Garnis C, Campbell J, Davies JJ, Macaulay C, Lam S, Lam WL. Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis. Hum Mol Genet. 2005; 14:475–482.

18. Pollack JR, S⊘ rlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002; 99:12963–12968.

19. Davies JJ, Wilson IM, Lam WL. Array CGH technologies and their applications to cancer genomes. Chromosome Res. 2005; 13:237–248.

20. Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genomewide measurement of DNA copy number. Nat Genet. 2001; 29:263–264.

21. Snijders AM, Pinkel D, Albertson DG. Current status and future prospects of array-based comparative genomic hybridisation. Brief Funct Genomic Proteomic. 2003; 2:37–45.

22. Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005; 6:331–654.

23. Albertson DG, Ylstra B, Segraves R, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000; 25:144–146.

24. Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004; 14:287–295.

25. Zhao X, Li C, Paez JG, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004; 64:3060–3071.

26. Kennedy GC, Matsuzaki H, Dong S, et al. Large-scale geno-typing of complex DNA. Nat Biotechnol. 2003; 21:1233–1237.

27. Lucito R, Healy J, Alexander J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003; 13:2291–2305.

28. Brennan C, Zhang Y, Leo C, et al. High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Res. 2004; 64:4744–4748.

29. Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004; 36:949–951.

30. Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004; 305:525–528.

31. Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004; 36:299–303.

32. Bruder CE, Hirvelä C, Tapia-Paez I, et al. High resolution deletion analysis of constitutional DNA from neurofibro-matosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet. 2001; 10:271–282.

33. Vissers LE, de Vries BB, Osoegawa K, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003; 73:1261–1270.
34. Veltman JA, Schoenmakers EF, Eussen BH, et al. High- throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet. 2002; 70:1269–1276.
35. Weiss MM, Snijders AM, Kuipers EJ, et al. Determination of amplicon boundaries at 20q13.2 in tissue samples of human gastric adenocarcinomas by high-resolution microarray comparative genomic hybridization. J Pathol. 2003; 200:320–326.

36. Carvalho B, Ouwerkerk E, Meijer GA, Ylstra B. High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J Clin Pathol. 2004; 57:644–646.

37. Shaw CJ, Stankiewicz P, Bien-Willner G, et al. Small marker chromosomes in two patients with segmental aneusomy for proximal 17p. Hum Genet. 2004; 115:1–7.
38. Buckley PG, Mantripragada KK, Benetkiewicz M, et al. A full-coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum Mol Genet. 2002; 11:3221–3229.

40. Waldman FM, DeVries S, Chew KL, et al. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J Natl Cancer Inst. 2000; 92:313–320.

41. Chin K, de Solorzano CO, Knowles D, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004; 36:984–988.

42. Cho EK, Tchinda J, Freeman JL, Chung YJ, Cai WW, Lee C. Array-based comparative genomic hybridization and copy number variation in cancer research. Cytogenet Genome Res. 2006; 115:262–272.

43. Yao J, Weremowicz S, Feng B, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006; 66:4065–4078.

44. Lai LA, Paulson TG, Li X, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007; 46:532–542.

46. Martinez-Climent JA, Alizadeh AA, Segraves R, et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003; 101:3109–3117.

47. Lossos IS, Alizadeh AA, Diehn M, et al. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alterna-tive patterns with increased or decreased expression of c-myc and its regulated genes. Proc Natl Acad Sci USA. 2002; 99:8886–8891.

48. Schwaenen C, Nessling M, Wessendorf S, et al. Automated array-based genomic profiling in chronic lymphocytic leuke-mia: development of a clinical tool and discovery of recurrent genomic alterations. Proc Natl Acad Sci USA. 2004; 101:1039–1044.

49. Weiss MM, Kuipers EJ, Postma C, et al. Genome wide array comparative genomic hybridisation analysis of premalignant lesions of the stomach. Mol Pathol. 2003; 56:293–298.

50. Paris PL, Andaya A, Fridlyand J, et al. Whole genome scann-ing identifies genotypes associated with recurrence and meta-stasis in prostate tumors. Hum Mol Genet. 2004; 13:1303–1313.

51. Callagy G, Pharoah P, Chin SF, et al. Identification and validation of prognostic markers in breast cancer with the complementary use of array-CGH and tissue microarrays. J Pathol. 2005; 205:388–396.

52. Nyante SJ, Devries S, Chen YY, Hwang ES. Array-based comparative genomic hybridization of ductal carcinoma in situ and synchronous invasive lobular cancer. Hum Patho. 2004; 35:759–763.

53. Costa JL, Meijer G, Ylstra B, Caldas C. Array comparative genomic hybridization copy number profiling: a new tool for translational research in solid malignancies. Semin Radiat Oncol. 2008; 18:98–104.

54. Birrer MJ, Johnson ME, Hao K, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007; 25:2281–2287.

55. Hirasaki S, Noguchi T, Mimori K, et al. BAC clones related to prognosis in patients with esophageal squamous carcinoma: an array comparative genomic hybridization study. Oncologist. 2007; 12:406–417.

56. Weiss MM, Kuipers EJ, Postma C, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004; 26:307–317.

57. Rubio-Moscardo F, Climent J, Siebert R, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005; 105:4445–4454.

58. Blaveri E, Brewer JL, Roydasgupta R, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005; 11:7012–7022.

59. Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophy-siologies. Cancer Cell. 2006; 10:529–541.

60. Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006; 45:1033–1040.

61. Chin SF, Wang Y, Thorne NP, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007; 26:1959–1970.

62. Kim SW, Kim JW, Kim YT, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007; 46:1–9.

63. Han W, Han MR, Kang JJ, et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006; 6:92.

64. Heidenblad M, Lindgren D, Veltman JA, et al. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene. 2005; 24:1794–1801.

65. Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003; 95:1878–1890.

66. Zardo G, Tiirikainen MI, Hong C, et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet. 2002; 32:453–458.

67. Zhou X, Mok SC, Chen Z, Li Y, Wong DT. Concurrent analysis of loss of heterozygosity (LOH) and copy number abnormality (CNA) for oral premalignancy progression using the Affymetrix 10K SNP mapping array. Hum Genet. 2004; 115:327–330.

68. Yu W, Ballif BC, Kashork CD, et al. Development of a comparative genomic hybridization microarray and demon-stration of its utility with 25 well-characterized 1p36 deletions. Hum Mol Genet. 2003; 12:2145–2152.

69. Locke DP, Segraves R, Nicholls RD, et al. BAC microarray analysis of 15q11-q13 rearrangements and the impact of segmental duplications. J Med Genet. 2004; 41:175–182.

70. Shaw-Smith C, Redon R, Rickman L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmor-phic features. J Med Genet. 2004; 41:241–248.

71. Veltman JA, Jonkers Y, Nuijten I, et al. Definition of a critical region on chromosome 18 for congenital aural atresia by arrayCGH. Am J Hum Genet. 2003; 72:1578–1584.

72. Rauen KA, Albertson DG, Pinkel D, Cotter PD. Additional patient with del(12)(q21.2q22): further evidence for a candidate region for cardio-facio-cutaneous syndrome? Am J Med Genet. 2002; 110:51–56.

73. Lockwood WW, Chari R, Chi B, Lam WL. Recent advances in array comparative genomic hybridization technologies and their applications in human genetics. Eur J Hum Genet. 2006; 14:139–148.

74. Greshock J, Naylor TL, Margolin A, et al. 1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis. Genome Res. 2004; 14:179–187.

75. Bejjani BA, Saleki R, Ballif BC, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more? Am J Med Genet A. 2005; 134:259–267.

76. Cheung SW, Shaw CA, Yu W, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005; 7:422–432.

77. Wilton L, Williamson R, McBain J, Edgar D, Voullaire L. Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N Engl J Med. 2001; 345:1537–1541.

78. Schaeffer AJ, Chung J, Heretis K, Wong A, Ledbetter DH, Lese Martin C. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet. 2004; 74:1168–1174.

79. Larrabee PB, Johnson KL, Pestova E, et al. Microarray analysis of cell-free fetal DNA in amniotic fluid: a prenatal molecular karyotype. Am J Hum Genet. 2004; 75:485–491.

Go to : 
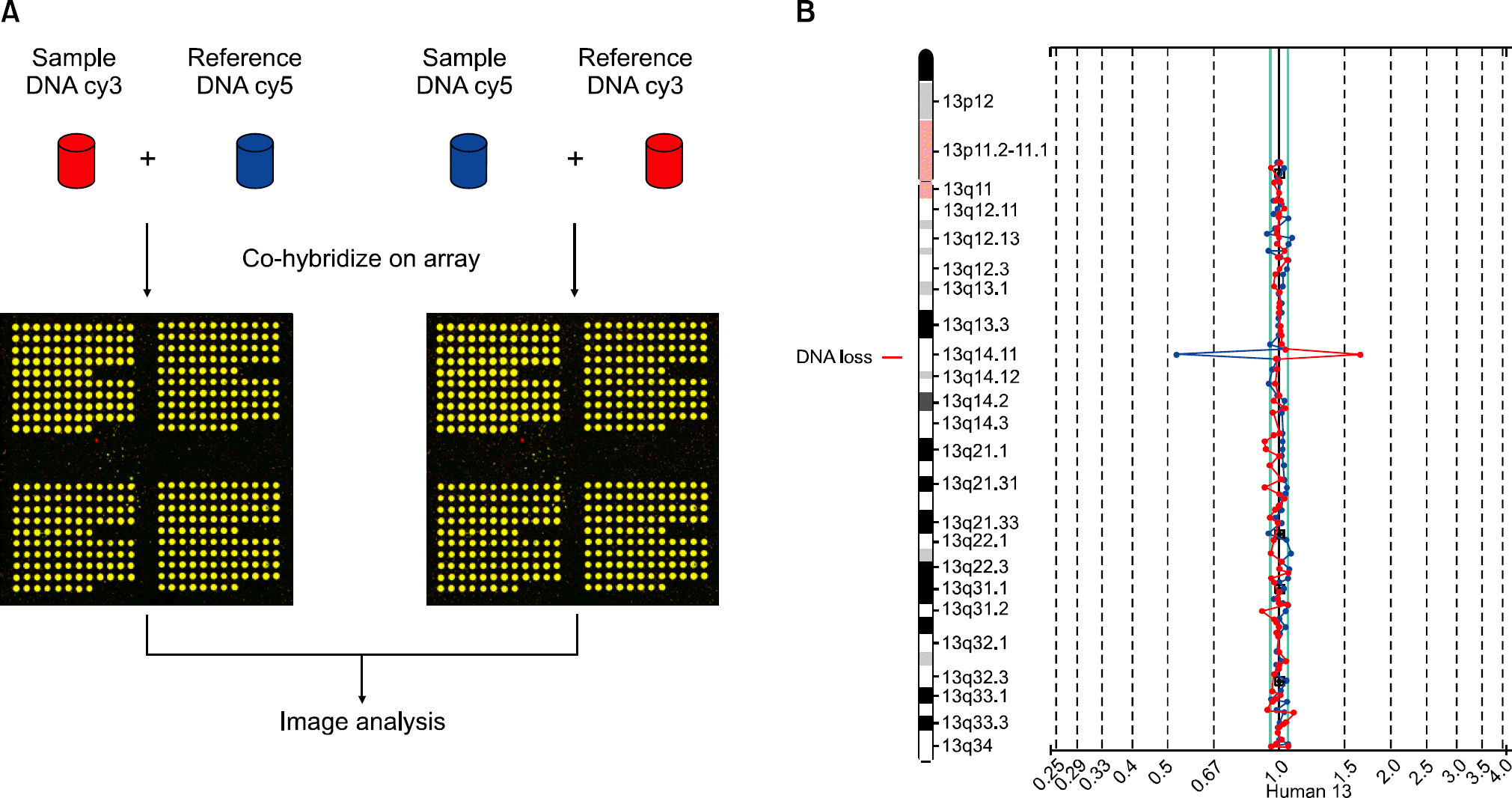 | Fig. 1.Array comparative genomic hybridization. (A) Sample and reference DNAs are differentially labeled with fluorescent dyes (typically, cyanine-3 and cyanine-5), combined, and co-hybridized to cloned DNA fragments, which are spotted on a glass slide. The sample and reference competitively bind to the spots and fluorescence intensity ratios resulting from hybridization of both DNAs are reflected by their relative quantities. (B) To reduce the false-positive error rate, the two profiles of a dye-swap experiment are compared. Data are normalized so that the ratio is set to some standard value, typically 1.0 on a linear scale or 0.0 on a logarithmic scale. Each dot on the graph represents a clone spotted on the array. Blue values to the left and red values to the right of the ‘1’ line indicate a loss of a genomic region, blue values to the right and red values to the left indicate a gain or amplification, and blue and red values at ‘1’ indicate no copy number change. A value of 0.5 as seen in this figure indicates a homozygous deletion. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download